Profile of Histone H3 Lysine 4 Trimethylation and the Effect of Lipopolysaccharide/Immune Complex-Activated Macrophages on Endotoxemia
- PMID: 31998290
- PMCID: PMC6965496
- DOI: 10.3389/fimmu.2019.02956
Profile of Histone H3 Lysine 4 Trimethylation and the Effect of Lipopolysaccharide/Immune Complex-Activated Macrophages on Endotoxemia
Abstract
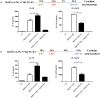
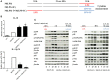
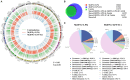
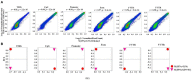
Macrophage plasticity is a process that allows macrophages to switch between two opposing phenotypes based on differential stimuli. Interferon γ (IFNγ)-primed macrophages stimulated with lipopolysaccharide (LPS) [M(IFNγ+LPS)] produce high levels of pro-inflammatory cytokines such as IL-12, TNFα, and IL-6 and low levels of the anti-inflammatory cytokine IL-10, while those stimulated with LPS in the presence of the immune complex (IC) [M(IFNγ+LPS+IC)] produce high levels of IL-10 and low levels of IL-12. In this study, we investigated the plasticity between M(IFNγ+LPS) and M(IFNγ+LPS+IC) in vitro and compared one of the active histone marks [histone H3 lysine 4 trimethylation (H3K4me3)] between M(IFNγ+LPS) and M(IFNγ+LPS+IC) using murine bone marrow-derived macrophages. We found that in an in vitro system, macrophages exhibited functional plasticity from M(LPS) to M(LPS+IC) upon repolarization after 2 days of washout period while IFNγ priming before LPS stimulation prevented this repolarization. Phosphorylation of p38, SAPK/JNK, and NF-κB p65 in M(LPS+IC) repolarized from M(LPS) was similar to that in M(LPS+IC) polarized from resting macrophages. To obtain the epigenetic profiles of M(IFNγ+LPS) and M(IFNγ+LPS+IC), the global enrichment of H3K4me3 was evaluated. M(IFNγ+LPS) and M(IFNγ+LPS+IC) displayed marked differences in genome-wide enrichment of H3K4me3. M(IFNγ+LPS+IC) showed increased global enrichment of H3K4me3, whereas M(IFNγ+LPS) showed decreased enrichment when compared to unstimulated macrophages. Furthermore, M(IFNγ+LPS+IC) exhibited high levels of H3K4me3 enrichment in all cis-regulatory elements. At the individual gene level, the results showed increased H3K4me3 enrichment in the promoters of known genes associated with M(IFNγ+LPS+IC), including Il10, Cxcl1, Csf3, and Il33, when compared with those of M(IFNγ+LPS). Finally, we investigated the impact of M(IFNγ+LPS+IC) on the systemic immune response by adoptive transfer of M(IFNγ+LPS+IC) in an LPS-induced endotoxemia model. The cytokine profile revealed that mice with adoptively transferred M(IFNγ+LPS+IC) had acutely reduced serum levels of the inflammatory cytokines IL-1β and IL-p12p70. This study highlights the importance of epigenetics in regulating macrophage activation and the functions of M(IFNγ+LPS+IC) that may influence macrophage plasticity and the potential therapeutic use of macrophage transfer in vivo.
Keywords: H3K4me3; LPS; endotoxemia; epigenetics; immune complex; macrophage.
Copyright © 2020 Ruenjaiman, Butta, Leu, Pongpanich, Leelahavanichkul, Kueanjinda and Palaga.
Figures


Similar articles
-
Notch signaling regulates the responses of lipopolysaccharide-stimulated macrophages in the presence of immune complexes.PLoS One. 2018 Jun 11;13(6):e0198609. doi: 10.1371/journal.pone.0198609. eCollection 2018. PLoS One. 2018. PMID: 29889863 Free PMC article.
-
Cytokine/Chemokine Release Patterns and Transcriptomic Profiles of LPS/IFNγ-Activated Human Macrophages Differentiated with Heat-Killed Mycobacterium obuense, M-CSF, or GM-CSF.Int J Mol Sci. 2021 Jul 5;22(13):7214. doi: 10.3390/ijms22137214. Int J Mol Sci. 2021. PMID: 34281268 Free PMC article.
-
The novel methyltransferase SETD4 regulates TLR agonist-induced expression of cytokines through methylation of lysine 4 at histone 3 in macrophages.Mol Immunol. 2019 Oct;114:179-188. doi: 10.1016/j.molimm.2019.07.011. Epub 2019 Jul 31. Mol Immunol. 2019. PMID: 31376731
-
Epigenetics and sex differences in the brain: A genome-wide comparison of histone-3 lysine-4 trimethylation (H3K4me3) in male and female mice.Exp Neurol. 2015 Jun;268:21-9. doi: 10.1016/j.expneurol.2014.08.006. Epub 2014 Aug 14. Exp Neurol. 2015. PMID: 25131640 Free PMC article. Review.
-
Epigenetics in susceptibility, progression, and diagnosis of periodontitis.Jpn Dent Sci Rev. 2022 Nov;58:183-192. doi: 10.1016/j.jdsr.2022.06.001. Epub 2022 Jun 17. Jpn Dent Sci Rev. 2022. PMID: 35754944 Free PMC article. Review.
Cited by
-
Epigenomic and transcriptomic analyses reveal differences between low-grade inflammation and severe exhaustion in LPS-challenged murine monocytes.Commun Biol. 2022 Jan 28;5(1):102. doi: 10.1038/s42003-022-03035-2. Commun Biol. 2022. PMID: 35091696 Free PMC article.
-
Less Severe Lipopolysaccharide-Induced Inflammation in Conditional mgmt-Deleted Mice with LysM-Cre System: The Loss of DNA Repair in Macrophages.Int J Mol Sci. 2023 Jun 14;24(12):10139. doi: 10.3390/ijms241210139. Int J Mol Sci. 2023. PMID: 37373287 Free PMC article.
-
Transmembrane Protein-Based Risk Model and H3K4me3 Modification Characteristics in Lung Adenocarcinoma.Front Oncol. 2022 Mar 22;12:828814. doi: 10.3389/fonc.2022.828814. eCollection 2022. Front Oncol. 2022. PMID: 35392225 Free PMC article.
-
The Regulatory Roles of Ezh2 in Response to Lipopolysaccharide (LPS) in Macrophages and Mice with Conditional Ezh2 Deletion with LysM-Cre System.Int J Mol Sci. 2023 Mar 10;24(6):5363. doi: 10.3390/ijms24065363. Int J Mol Sci. 2023. PMID: 36982437 Free PMC article.
-
Less Severe Polymicrobial Sepsis in Conditional mgmt-Deleted Mice Using LysM-Cre System, Impacts of DNA Methylation and MGMT Inhibitor in Sepsis.Int J Mol Sci. 2023 Jun 15;24(12):10175. doi: 10.3390/ijms241210175. Int J Mol Sci. 2023. PMID: 37373325 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

