Multiplex Neural Circuit Tracing With G-Deleted Rabies Viral Vectors
- PMID: 31998081
- PMCID: PMC6967742
- DOI: 10.3389/fncir.2019.00077
Multiplex Neural Circuit Tracing With G-Deleted Rabies Viral Vectors
Abstract
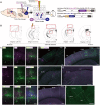
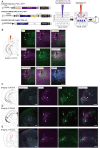
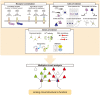
Neural circuits interconnect to organize large-scale networks that generate perception, cognition, memory, and behavior. Information in the nervous system is processed both through parallel, independent circuits and through intermixing circuits. Analyzing the interaction between circuits is particularly indispensable for elucidating how the brain functions. Monosynaptic circuit tracing with glycoprotein (G) gene-deleted rabies viral vectors (RVΔG) comprises a powerful approach for studying the structure and function of neural circuits. Pseudotyping of RVΔG with the foreign envelope EnvA permits expression of transgenes such as fluorescent proteins, genetically-encoded sensors, or optogenetic tools in cells expressing TVA, a cognate receptor for EnvA. Trans-complementation with rabies virus glycoproteins (RV-G) enables trans-synaptic labeling of input neurons directly connected to the starter neurons expressing both TVA and RV-G. However, it remains challenging to simultaneously map neuronal connections from multiple cell populations and their interactions between intermixing circuits solely with the EnvA/TVA-mediated RV tracing system in a single animal. To overcome this limitation, here, we multiplexed RVΔG circuit tracing by optimizing distinct viral envelopes (oEnvX) and their corresponding receptors (oTVX). Based on the EnvB/TVB and EnvE/DR46-TVB systems derived from the avian sarcoma leukosis virus (ASLV), we developed optimized TVB receptors with lower or higher affinity (oTVB-L or oTVB-H) and the chimeric envelope oEnvB, as well as an optimized TVE receptor with higher affinity (oTVE-H) and its chimeric envelope oEnvE. We demonstrated independence of RVΔG infection between the oEnvA/oTVA, oEnvB/oTVB, and oEnvE/oTVE systems and in vivo proof-of-concept for multiplex circuit tracing from two distinct classes of layer 5 neurons targeting either other cortical or subcortical areas. We also successfully labeled common input of the lateral geniculate nucleus to both cortico-cortical layer 5 neurons and inhibitory neurons of the mouse V1 with multiplex RVΔG tracing. These oEnvA/oTVA, oEnvB/oTVB, and oEnvE/oTVE systems allow for differential labeling of distinct circuits to uncover the mechanisms underlying parallel processing through independent circuits and integrated processing through interaction between circuits in the brain.
Keywords: anatomy; multiplex; neural circuit; rabies virus; transsynaptic targeting.
Copyright © 2020 Suzuki, Morimoto, Akaike and Osakada.
Figures

Similar articles
-
Monosynaptic rabies virus tracing from projection-targeted single neurons.Neurosci Res. 2022 May;178:20-32. doi: 10.1016/j.neures.2022.01.007. Epub 2022 Jan 31. Neurosci Res. 2022. PMID: 35101519
-
Identification of Two Classes of Somatosensory Neurons That Display Resistance to Retrograde Infection by Rabies Virus.J Neurosci. 2017 Oct 25;37(43):10358-10371. doi: 10.1523/JNEUROSCI.1277-17.2017. Epub 2017 Sep 26. J Neurosci. 2017. PMID: 28951448 Free PMC article.
-
Design and generation of recombinant rabies virus vectors.Nat Protoc. 2013 Aug;8(8):1583-601. doi: 10.1038/nprot.2013.094. Epub 2013 Jul 25. Nat Protoc. 2013. PMID: 23887178 Free PMC article.
-
G gene-deficient single-round rabies viruses for neuronal circuit analysis.Virus Res. 2016 May 2;216:41-54. doi: 10.1016/j.virusres.2015.05.023. Epub 2015 Jun 8. Virus Res. 2016. PMID: 26065596 Review.
-
Monosynaptic Circuit Tracing with Glycoprotein-Deleted Rabies Viruses.J Neurosci. 2015 Jun 17;35(24):8979-85. doi: 10.1523/JNEUROSCI.0409-15.2015. J Neurosci. 2015. PMID: 26085623 Free PMC article. Review. No abstract available.
Cited by
-
Violet light suppresses lens-induced myopia via neuropsin (OPN5) in mice.Proc Natl Acad Sci U S A. 2021 Jun 1;118(22):e2018840118. doi: 10.1073/pnas.2018840118. Proc Natl Acad Sci U S A. 2021. PMID: 34031241 Free PMC article.
-
Leveraging the replication-competent avian-like sarcoma virus/tumor virus receptor-A system for modeling human gliomas.Glia. 2021 Sep;69(9):2059-2076. doi: 10.1002/glia.23984. Epub 2021 Feb 27. Glia. 2021. PMID: 33638562 Free PMC article. Review.
-
Application of Recombinant Rabies Virus to Xenopus Tadpole Brain.eNeuro. 2021 Jun 7;8(4):ENEURO.0477-20.2021. doi: 10.1523/ENEURO.0477-20.2021. Online ahead of print. eNeuro. 2021. PMID: 34099488 Free PMC article.
-
In Vivo Whole-Cell Patch-Clamp Methods: Recent Technical Progress and Future Perspectives.Sensors (Basel). 2021 Feb 19;21(4):1448. doi: 10.3390/s21041448. Sensors (Basel). 2021. PMID: 33669656 Free PMC article. Review.
-
New rabies viral resources for multi-scale neural circuit mapping.Mol Psychiatry. 2024 Jul;29(7):1951-1967. doi: 10.1038/s41380-024-02451-6. Epub 2024 Feb 14. Mol Psychiatry. 2024. PMID: 38355784 Free PMC article.
References
-
- Adkins H. B., Brojatsch J., Young J. A. T. (2000). Identification and characterization of a shared TNFR-related receptor for subgroup B, D, and E avian leukosis viruses reveal cysteine residues required specifically for subgroup E viral entry. J. Virol. 74, 3572–3578. 10.1128/jvi.74.8.3572-3578.2000 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

