Accelerated brain aging towards transcriptional inversion in a zebrafish model of the K115fs mutation of human PSEN2
- PMID: 31978074
- PMCID: PMC6980398
- DOI: 10.1371/journal.pone.0227258
Accelerated brain aging towards transcriptional inversion in a zebrafish model of the K115fs mutation of human PSEN2
Abstract
Background: The molecular changes involved in Alzheimer's disease (AD) progression remain unclear since we cannot easily access antemortem human brains. Some non-mammalian vertebrates such as the zebrafish preserve AD-relevant transcript isoforms of the PRESENILIN genes lost from mice and rats. One example is PS2V, the alternative transcript isoform of the PSEN2 gene. PS2V is induced by hypoxia/oxidative stress and shows increased expression in late onset, sporadic AD brains. A unique, early onset familial AD mutation of PSEN2, K115fs, mimics the PS2V coding sequence suggesting that forced, early expression of PS2V-like isoforms may contribute to AD pathogenesis. Here we use zebrafish to model the K115fs mutation to investigate the effects of forced PS2V-like expression on the transcriptomes of young adult and aged adult brains.
Methods: We edited the zebrafish genome to model the K115fs mutation. To explore its effects at the molecular level, we analysed the brain transcriptome and proteome of young (6-month-old) and aged (24-month-old) wild type and heterozygous mutant female sibling zebrafish. Finally, we used gene co-expression network analysis (WGCNA) to compare molecular changes in the brains of these fish to human AD.
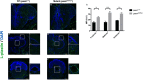
Results: Young heterozygous mutant fish show transcriptional changes suggesting accelerated brain aging and increased glucocorticoid signalling. These early changes precede a transcriptional 'inversion' that leads to glucocorticoid resistance and other likely pathological changes in aged heterozygous mutant fish. Notably, microglia-associated immune responses regulated by the ETS transcription factor family are altered in both our zebrafish mutant model and in human AD. The molecular changes we observe in aged heterozygous mutant fish occur without obvious histopathology and possibly in the absence of Aβ.
Conclusions: Our results suggest that forced expression of a PS2V-like isoform contributes to immune and stress responses favouring AD pathogenesis. This highlights the value of our zebrafish genetic model for exploring molecular mechanisms involved in AD pathogenesis.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Alzheimer's disease-related peptide PS2V plays ancient, conserved roles in suppression of the unfolded protein response under hypoxia and stimulation of γ-secretase activity.Hum Mol Genet. 2015 Jul 1;24(13):3662-78. doi: 10.1093/hmg/ddv110. Epub 2015 Mar 26. Hum Mol Genet. 2015. PMID: 25814654
-
The Zebrafish Equivalent of Alzheimer's Disease-Associated PRESENILIN Isoform PS2V Regulates Inflammatory and Other Responses to Hypoxic Stress.J Alzheimers Dis. 2016 Mar 31;52(2):581-608. doi: 10.3233/JAD-150678. J Alzheimers Dis. 2016. PMID: 27031468
-
Brain transcriptome analysis of a familial Alzheimer's disease-like mutation in the zebrafish presenilin 1 gene implies effects on energy production.Mol Brain. 2019 May 3;12(1):43. doi: 10.1186/s13041-019-0467-y. Mol Brain. 2019. PMID: 31053140 Free PMC article.
-
Exploring Alzheimer's disease in zebrafish.J Alzheimers Dis. 2010;20(4):981-90. doi: 10.3233/JAD-2010-1412. J Alzheimers Dis. 2010. PMID: 20182049 Review.
-
A Review of the Familial Alzheimer's Disease Locus PRESENILIN 2 and Its Relationship to PRESENILIN 1.J Alzheimers Dis. 2018;66(4):1323-1339. doi: 10.3233/JAD-180656. J Alzheimers Dis. 2018. PMID: 30412492 Review.
Cited by
-
From multiallele fish to nonstandard environments, how ZFIN assigns phenotypes, human disease models, and gene expression annotations to genes.Genetics. 2023 May 4;224(1):iyad032. doi: 10.1093/genetics/iyad032. Genetics. 2023. PMID: 36864549 Free PMC article.
-
Use of Zebrafish Genetic Models to Study Etiology of the Amyloid-Beta and Neurofibrillary Tangle Pathways in Alzheimer's Disease.Curr Neuropharmacol. 2022 Mar 4;20(3):524-539. doi: 10.2174/1570159X19666210524155944. Curr Neuropharmacol. 2022. PMID: 34030617 Free PMC article. Review.
-
Zebrafish Chromosome 14 Gene Differential Expression in the fmr1 h u2787 Model of Fragile X Syndrome.Front Genet. 2021 May 31;12:625466. doi: 10.3389/fgene.2021.625466. eCollection 2021. Front Genet. 2021. PMID: 34135935 Free PMC article.
-
Zebrafish as a Model Organism for Studying Pathologic Mechanisms of Neurodegenerative Diseases and other Neural Disorders.Cell Mol Neurobiol. 2023 Aug;43(6):2603-2620. doi: 10.1007/s10571-023-01340-w. Epub 2023 Apr 1. Cell Mol Neurobiol. 2023. PMID: 37004595 Free PMC article. Review.
-
Accelerated loss of hypoxia response in zebrafish with familial Alzheimer's disease-like mutation of presenilin 1.Hum Mol Genet. 2020 Aug 11;29(14):2379-2394. doi: 10.1093/hmg/ddaa119. Hum Mol Genet. 2020. PMID: 32588886 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

