PreS1 Mutations Alter the Large HBsAg Antigenicity of a Hepatitis B Virus Strain Isolated in Bangladesh
- PMID: 31952213
- PMCID: PMC7014173
- DOI: 10.3390/ijms21020546
PreS1 Mutations Alter the Large HBsAg Antigenicity of a Hepatitis B Virus Strain Isolated in Bangladesh
Abstract
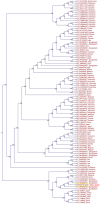
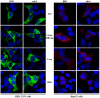
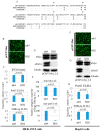
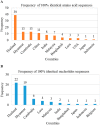
Mutations in the hepatitis B virus (HBV) genome can potentially lead to vaccination failure, diagnostic escape, and disease progression. However, there are no reports on viral gene expression and large hepatitis B surface antigen (HBsAg) antigenicity alterations due to mutations in HBV isolated from a Bangladeshi population. Here, we sequenced the full genome of the HBV isolated from a clinically infected patient in Bangladesh. The open reading frames (ORFs) (P, S, C, and X) of the isolated HBV strain were successfully amplified and cloned into a mammalian expression vector. The HBV isolate was identified as genotype C (sub-genotype C2), serotype adr, and evolutionarily related to strains isolated in Indonesia, Malaysia, and China. Clinically significant mutations, such as preS1 C2964A, reverse transcriptase domain I91L, and small HBsAg N3S, were identified. The viral P, S, C, and X genes were expressed in HEK-293T and HepG2 cells by transient transfection with a native subcellular distribution pattern analyzed by immunofluorescence assay. Western blotting of large HBsAg using preS1 antibody showed no staining, and preS1 ELISA showed a significant reduction in reactivity due to amino acid mutations. This mutated preS1 sequence has been identified in several Asian countries. To our knowledge, this is the first report investigating changes in large HBsAg antigenicity due to preS1 mutations.
Keywords: Bangladesh; antigenicity; full genome sequence; hepatitis B virus (HBV); preS1 mutations.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
Both middle and large envelope proteins can mediate neutralization of hepatitis B virus infectivity by anti-preS2 antibodies: escape by naturally occurring preS2 deletions.J Virol. 2024 Aug 20;98(8):e0192923. doi: 10.1128/jvi.01929-23. Epub 2024 Jul 30. J Virol. 2024. PMID: 39078152
-
A monoclonal antibody specific to the non-epitope region of hepatitis B virus preS1 contributes to more effective HBV detection.Clin Biochem. 2013 Aug;46(12):1105-1110. doi: 10.1016/j.clinbiochem.2013.04.011. Epub 2013 Apr 19. Clin Biochem. 2013. PMID: 23608352
-
Expression and immunoactivity of chimeric particulate antigens of receptor binding site-core antigen of hepatitis B virus.World J Gastroenterol. 2005 Jan 28;11(4):492-7. doi: 10.3748/wjg.v11.i4.492. World J Gastroenterol. 2005. PMID: 15641132 Free PMC article.
-
The virological and clinical significance of mutations in the overlapping envelope and polymerase genes of hepatitis B virus.J Clin Virol. 2002 Aug;25(2):97-106. doi: 10.1016/s1386-6532(02)00049-5. J Clin Virol. 2002. PMID: 12367644 Review.
-
Applications of human hepatitis B virus preS domain in bio- and nanotechnology.World J Gastroenterol. 2015 Jun 28;21(24):7400-11. doi: 10.3748/wjg.v21.i24.7400. World J Gastroenterol. 2015. PMID: 26139986 Free PMC article. Review.
Cited by
-
Complete Genome Sequence of a Precore-Defective Hepatitis B Virus Genotype D2 Strain Isolated in Bangladesh.Microbiol Resour Announc. 2020 Mar 12;9(11):e00083-20. doi: 10.1128/MRA.00083-20. Microbiol Resour Announc. 2020. PMID: 32165384 Free PMC article.
-
Computational Analysis Predicts Correlations among Amino Acids in SARS-CoV-2 Proteomes.Biomedicines. 2023 Feb 10;11(2):512. doi: 10.3390/biomedicines11020512. Biomedicines. 2023. PMID: 36831052 Free PMC article.
-
Characteristics of Hepatitis B Virus Genotype and Sub-Genotype in Hepatocellular Cancer Patients in Vietnam.Diagnostics (Basel). 2022 Oct 1;12(10):2393. doi: 10.3390/diagnostics12102393. Diagnostics (Basel). 2022. PMID: 36292082 Free PMC article.
-
Impact of the Interaction of Hepatitis B Virus with Mitochondria and Associated Proteins.Viruses. 2020 Feb 4;12(2):175. doi: 10.3390/v12020175. Viruses. 2020. PMID: 32033216 Free PMC article. Review.
-
Complete genome sequence of hepatitis B virus identified from a patient suffering from chronic kidney disease in Bangladesh.Microbiol Resour Announc. 2024 May 9;13(5):e0115123. doi: 10.1128/mra.01151-23. Epub 2024 Apr 16. Microbiol Resour Announc. 2024. PMID: 38624203 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

