Large-Scale Profiling of RBP-circRNA Interactions from Public CLIP-Seq Datasets
- PMID: 31947823
- PMCID: PMC7016857
- DOI: 10.3390/genes11010054
Large-Scale Profiling of RBP-circRNA Interactions from Public CLIP-Seq Datasets
Abstract
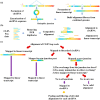
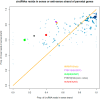
Circular RNAs are a special type of RNA that has recently attracted a lot of research interest in studying its formation and function. RNA binding proteins (RBPs) that bind circRNAs are important in these processes, but have been relatively less studied. CLIP-Seq technology has been invented and applied to profile RBP-RNA interactions on the genome-wide scale. While mRNAs are usually the focus of CLIP-Seq experiments, RBP-circRNA interactions could also be identified through specialized analysis of CLIP-Seq datasets. However, many technical difficulties are involved in this process, such as the usually short read length of CLIP-Seq reads. In this study, we created a pipeline called Clirc specialized for profiling circRNAs in CLIP-Seq data and analyzing the characteristics of RBP-circRNA interactions. In conclusion, to our knowledge, this is one of the first studies to investigate circRNAs and their binding partners through repurposing CLIP-Seq datasets, and we hope our work will become a valuable resource for future studies into the biogenesis and function of circRNAs.
Keywords: CLIP-Seq; Circ-RNA; RBP.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Mapping the Transcriptome-Wide Landscape of RBP Binding Sites Using gPAR-CLIP-seq: Bioinformatic Analysis.Methods Mol Biol. 2016;1361:91-104. doi: 10.1007/978-1-4939-3079-1_6. Methods Mol Biol. 2016. PMID: 26483018
-
CLIPdb: a CLIP-seq database for protein-RNA interactions.BMC Genomics. 2015 Feb 5;16(1):51. doi: 10.1186/s12864-015-1273-2. BMC Genomics. 2015. PMID: 25652745 Free PMC article.
-
Transcriptome-wide profiles of circular RNA and RNA-binding protein interactions reveal effects on circular RNA biogenesis and cancer pathway expression.Genome Med. 2020 Dec 7;12(1):112. doi: 10.1186/s13073-020-00812-8. Genome Med. 2020. PMID: 33287884 Free PMC article.
-
Computational analysis of CLIP-seq data.Methods. 2017 Apr 15;118-119:60-72. doi: 10.1016/j.ymeth.2017.02.006. Epub 2017 Feb 22. Methods. 2017. PMID: 28254606 Review.
-
Practical considerations on performing and analyzing CLIP-seq experiments to identify transcriptomic-wide RNA-protein interactions.Methods. 2019 Feb 15;155:49-57. doi: 10.1016/j.ymeth.2018.12.002. Epub 2018 Dec 6. Methods. 2019. PMID: 30527764 Free PMC article. Review.
Cited by
-
The Biomarker and Therapeutic Potential of Circular Rnas in Schizophrenia.Cells. 2020 Oct 4;9(10):2238. doi: 10.3390/cells9102238. Cells. 2020. PMID: 33020462 Free PMC article. Review.
-
Establishment and verification of a prognostic model of liver cancer by RNA-binding proteins based on the TCGA database.Transl Cancer Res. 2022 Jul;11(7):1925-1937. doi: 10.21037/tcr-21-2820. Transl Cancer Res. 2022. PMID: 36249884 Free PMC article.
-
FL-circAS: an integrative resource and analysis for full-length sequences and alternative splicing of circular RNAs with nanopore sequencing.Nucleic Acids Res. 2024 Jan 5;52(D1):D115-D123. doi: 10.1093/nar/gkad829. Nucleic Acids Res. 2024. PMID: 37823705 Free PMC article.
-
Evaluation of methods to detect circular RNAs from single-end RNA-sequencing data.BMC Genomics. 2022 Feb 8;23(1):106. doi: 10.1186/s12864-022-08329-7. BMC Genomics. 2022. PMID: 35135477 Free PMC article.
-
Exosome-Associated circRNAs as Key Regulators of EMT in Cancer.Cells. 2022 May 23;11(10):1716. doi: 10.3390/cells11101716. Cells. 2022. PMID: 35626752 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

