Energy Metabolism in Cancer: The Roles of STAT3 and STAT5 in the Regulation of Metabolism-Related Genes
- PMID: 31947710
- PMCID: PMC7016889
- DOI: 10.3390/cancers12010124
Energy Metabolism in Cancer: The Roles of STAT3 and STAT5 in the Regulation of Metabolism-Related Genes
Abstract
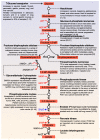
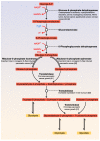
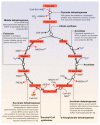
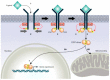
A central characteristic of many types of cancer is altered energy metabolism processes such as enhanced glucose uptake and glycolysis and decreased oxidative metabolism. The regulation of energy metabolism is an elaborate process involving regulatory proteins such as HIF (pro-metastatic protein), which reduces oxidative metabolism, and some other proteins such as tumour suppressors that promote oxidative phosphorylation. In recent years, it has been demonstrated that signal transducer and activator of transcription (STAT) proteins play a pivotal role in metabolism regulation. STAT3 and STAT5 are essential regulators of cytokine- or growth factor-induced cell survival and proliferation, as well as the crosstalk between STAT signalling and oxidative metabolism. Several reports suggest that the constitutive activation of STAT proteins promotes glycolysis through the transcriptional activation of hypoxia-inducible factors and therefore, the alteration of mitochondrial activity. It seems that STAT proteins function as an integrative centre for different growth and survival signals for energy and respiratory metabolism. This review summarises the functions of STAT3 and STAT5 in the regulation of some metabolism-related genes and the importance of oxygen in the tumour microenvironment to regulate cell metabolism, particularly in the metabolic pathways that are involved in energy production in cancer cells.
Keywords: HIF; STAT3; STAT5; Warburg effect; cancer metabolism; transcription factors.
Conflict of interest statement
The authors report no conflict of interest. The authors alone are responsible for the content and writing of this manuscript.
Figures

Similar articles
-
STAT proteins in cancer: orchestration of metabolism.Nat Rev Cancer. 2023 Mar;23(3):115-134. doi: 10.1038/s41568-022-00537-3. Epub 2023 Jan 3. Nat Rev Cancer. 2023. PMID: 36596870 Review.
-
STAT proteins as novel targets for cancer drug discovery.Expert Opin Ther Targets. 2004 Oct;8(5):409-22. doi: 10.1517/14728222.8.5.409. Expert Opin Ther Targets. 2004. PMID: 15469392 Review.
-
STAT5 Is Necessary for the Metabolic Switch Induced by IL-2 in Cervical Cancer Cell Line SiHa.Int J Mol Sci. 2024 Jun 21;25(13):6835. doi: 10.3390/ijms25136835. Int J Mol Sci. 2024. PMID: 38999946 Free PMC article.
-
Involvement of STAT5 in Oncogenesis.Biomedicines. 2020 Aug 28;8(9):316. doi: 10.3390/biomedicines8090316. Biomedicines. 2020. PMID: 32872372 Free PMC article. Review.
-
The genes induced by signal transducer and activators of transcription (STAT)3 and STAT5 in mammary epithelial cells define the roles of these STATs in mammary development.Mol Endocrinol. 2006 Mar;20(3):675-85. doi: 10.1210/me.2005-0392. Epub 2005 Nov 17. Mol Endocrinol. 2006. PMID: 16293640
Cited by
-
WZB117 enhanced the anti-tumor effect of apatinib against melanoma via blocking STAT3/PKM2 axis.Front Pharmacol. 2022 Sep 16;13:976117. doi: 10.3389/fphar.2022.976117. eCollection 2022. Front Pharmacol. 2022. PMID: 36188586 Free PMC article.
-
Metformin: Metabolic Rewiring Faces Tumor Heterogeneity.Cells. 2020 Nov 9;9(11):2439. doi: 10.3390/cells9112439. Cells. 2020. PMID: 33182253 Free PMC article. Review.
-
Keratinocyte-induced costimulation of human T cells through CD6 - but not CD2 - activates mTOR and prevents oxidative stress.Front Immunol. 2022 Oct 24;13:1016112. doi: 10.3389/fimmu.2022.1016112. eCollection 2022. Front Immunol. 2022. PMID: 36353616 Free PMC article.
-
Regulation of Metabolic Plasticity in Cancer Stem Cells and Implications in Cancer Therapy.Cancers (Basel). 2022 Nov 30;14(23):5912. doi: 10.3390/cancers14235912. Cancers (Basel). 2022. PMID: 36497394 Free PMC article. Review.
-
Targeting the complement system in pancreatic cancer drug resistance: a novel therapeutic approach.Cancer Drug Resist. 2022 Apr 3;5(2):317-327. doi: 10.20517/cdr.2021.150. eCollection 2022. Cancer Drug Resist. 2022. PMID: 35800364 Free PMC article. Review.
References
-
- Younes M., Brown R.W., Mody D.R., Fernandez L., Laucirica R. GLUT1 expression in human breast carcinoma: Correlation with known prognostic markers. Anticancer Res. 1995;15:2895–2898. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

