A Tale of Two Viruses: The Distinct Spike Glycoproteins of Feline Coronaviruses
- PMID: 31936749
- PMCID: PMC7019228
- DOI: 10.3390/v12010083
A Tale of Two Viruses: The Distinct Spike Glycoproteins of Feline Coronaviruses
Abstract
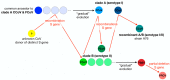
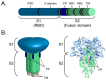
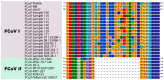
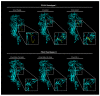
Feline coronavirus (FCoV) is a complex viral agent that causes a variety of clinical manifestations in cats, commonly known as feline infectious peritonitis (FIP). It is recognized that FCoV can occur in two different serotypes. However, differences in the S protein are much more than serological or antigenic variants, resulting in the effective presence of two distinct viruses. Here, we review the distinct differences in the S proteins of these viruses, which are likely to translate into distinct biological outcomes. We introduce a new concept related to the non-taxonomical classification and differentiation among FCoVs by analyzing and comparing the genetic, structural, and functional characteristics of FCoV and the FCoV S protein among the two serotypes and FCoV biotypes. Based on our analysis, we suggest that our understanding of FIP needs to consider whether the presence of these two distinct viruses has implications in clinical settings.
Keywords: feline coronavirus; feline infectious peritonitis; genetic characterization; serotype; spike protein.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Feline Coronaviruses: Pathogenesis of Feline Infectious Peritonitis.Adv Virus Res. 2016;96:193-218. doi: 10.1016/bs.aivir.2016.08.002. Epub 2016 Aug 31. Adv Virus Res. 2016. PMID: 27712624 Free PMC article. Review.
-
Feline coronavirus: Insights into viral pathogenesis based on the spike protein structure and function.Virology. 2018 Apr;517:108-121. doi: 10.1016/j.virol.2017.12.027. Epub 2018 Jan 10. Virology. 2018. PMID: 29329682 Free PMC article.
-
Differential susceptibility of macrophages to serotype II feline coronaviruses correlates with differences in the viral spike protein.Virus Res. 2018 Aug 15;255:14-23. doi: 10.1016/j.virusres.2018.06.010. Epub 2018 Jun 21. Virus Res. 2018. PMID: 29936068 Free PMC article.
-
Circulation and genetic diversity of Feline coronavirus type I and II from clinically healthy and FIP-suspected cats in China.Transbound Emerg Dis. 2019 Mar;66(2):763-775. doi: 10.1111/tbed.13081. Epub 2018 Dec 5. Transbound Emerg Dis. 2019. PMID: 30468573 Free PMC article.
-
Genetic determinants of pathogenesis by feline infectious peritonitis virus.Vet Immunol Immunopathol. 2011 Oct 15;143(3-4):265-8. doi: 10.1016/j.vetimm.2011.06.021. Epub 2011 Jun 12. Vet Immunol Immunopathol. 2011. PMID: 21719115 Free PMC article. Review.
Cited by
-
Genetic and Phylogenetic Analysis of Feline Coronavirus in Guangxi Province of China from 2021 to 2024.Vet Sci. 2024 Sep 25;11(10):455. doi: 10.3390/vetsci11100455. Vet Sci. 2024. PMID: 39453047 Free PMC article.
-
Serologic, Virologic and Pathologic Features of Cats with Naturally Occurring Feline Infectious Peritonitis Enrolled in Antiviral Clinical Trials.Viruses. 2024 Mar 17;16(3):462. doi: 10.3390/v16030462. Viruses. 2024. PMID: 38543827 Free PMC article.
-
Detection and characterization of novel luchacoviruses, genus Alphacoronavirus, in saliva and feces of meso-carnivores in the northeastern United States.J Virol. 2023 Nov 30;97(11):e0082923. doi: 10.1128/jvi.00829-23. Epub 2023 Oct 26. J Virol. 2023. PMID: 37882520 Free PMC article.
-
Feline Infectious Peritonitis: European Advisory Board on Cat Diseases Guidelines.Viruses. 2023 Aug 31;15(9):1847. doi: 10.3390/v15091847. Viruses. 2023. PMID: 37766254 Free PMC article. Review.
-
Superior possibilities and upcoming horizons for nanoscience in COVID-19: noteworthy approach for effective diagnostics and management of SARS-CoV-2 outbreak.Chem Zvesti. 2023 Apr 4:1-24. doi: 10.1007/s11696-023-02795-3. Online ahead of print. Chem Zvesti. 2023. PMID: 37362791 Free PMC article. Review.
References
-
- Masters P.S., Perlman S. Coronaviridae. In: Knipe D.M., Howley P.M., editors. Fields Virology. 6th ed. Vol. 1. Lippincot Williams & Wilkins; Philadelphia, PA, USA: 2013. pp. 825–858.
-
- Whittaker G.R. Coronaviridae. In: MacLachlan N.J., Dubovi E.J., editors. Fenner’s Veterinary Virology. 5th ed. Academic Press; London, UK: 2017. pp. 435–462.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

