Resolvases, Dissolvases, and Helicases in Homologous Recombination: Clearing the Road for Chromosome Segregation
- PMID: 31936378
- PMCID: PMC7017083
- DOI: 10.3390/genes11010071
Resolvases, Dissolvases, and Helicases in Homologous Recombination: Clearing the Road for Chromosome Segregation
Abstract
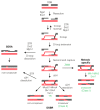
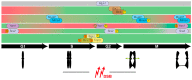
The execution of recombinational pathways during the repair of certain DNA lesions or in the meiotic program is associated to the formation of joint molecules that physically hold chromosomes together. These structures must be disengaged prior to the onset of chromosome segregation. Failure in the resolution of these linkages can lead to chromosome breakage and nondisjunction events that can alter the normal distribution of the genomic material to the progeny. To avoid this situation, cells have developed an arsenal of molecular complexes involving helicases, resolvases, and dissolvases that recognize and eliminate chromosome links. The correct orchestration of these enzymes promotes the timely removal of chromosomal connections ensuring the efficient segregation of the genome during cell division. In this review, we focus on the role of different DNA processing enzymes that collaborate in removing the linkages generated during the activation of the homologous recombination machinery as a consequence of the appearance of DNA breaks during the mitotic and meiotic programs. We will also discuss about the temporal regulation of these factors along the cell cycle, the consequences of their loss of function, and their specific role in the removal of chromosomal links to ensure the accurate segregation of the genomic material during cell division.
Keywords: DSB repair; dissolvases; helicases; homologous recombination; meiosis; mitosis; resolvases.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures

Similar articles
-
Alignment of Homologous Chromosomes and Effective Repair of Programmed DNA Double-Strand Breaks during Mouse Meiosis Require the Minichromosome Maintenance Domain Containing 2 (MCMDC2) Protein.PLoS Genet. 2016 Oct 19;12(10):e1006393. doi: 10.1371/journal.pgen.1006393. eCollection 2016 Oct. PLoS Genet. 2016. PMID: 27760146 Free PMC article.
-
Dbl2 Regulates Rad51 and DNA Joint Molecule Metabolism to Ensure Proper Meiotic Chromosome Segregation.PLoS Genet. 2016 Jun 15;12(6):e1006102. doi: 10.1371/journal.pgen.1006102. eCollection 2016 Jun. PLoS Genet. 2016. PMID: 27304859 Free PMC article.
-
The spatial regulation of meiotic recombination hotspots: are all DSB hotspots crossover hotspots?Exp Cell Res. 2012 Jul 15;318(12):1347-52. doi: 10.1016/j.yexcr.2012.03.025. Epub 2012 Mar 31. Exp Cell Res. 2012. PMID: 22487095 Review.
-
Self-organization of meiotic recombination initiation: general principles and molecular pathways.Annu Rev Genet. 2014;48:187-214. doi: 10.1146/annurev-genet-120213-092304. Annu Rev Genet. 2014. PMID: 25421598 Free PMC article. Review.
-
Spore-autonomous fluorescent protein expression identifies meiotic chromosome mis-segregation as the principal cause of hybrid sterility in yeast.PLoS Biol. 2018 Nov 12;16(11):e2005066. doi: 10.1371/journal.pbio.2005066. eCollection 2018 Nov. PLoS Biol. 2018. PMID: 30419022 Free PMC article.
Cited by
-
SWR1-Independent Association of H2A.Z to the LINC Complex Promotes Meiotic Chromosome Motion.Front Cell Dev Biol. 2020 Oct 22;8:594092. doi: 10.3389/fcell.2020.594092. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 33195270 Free PMC article.
-
The Cdc14 Phosphatase Controls Resolution of Recombination Intermediates and Crossover Formation during Meiosis.Int J Mol Sci. 2021 Sep 10;22(18):9811. doi: 10.3390/ijms22189811. Int J Mol Sci. 2021. PMID: 34575966 Free PMC article.
-
Deletion of IRC19 Causes Defects in DNA Double-Strand Break Repair Pathways in Saccharomyces cerevisiae.J Microbiol. 2024 Sep;62(9):749-758. doi: 10.1007/s12275-024-00152-x. Epub 2024 Jul 12. J Microbiol. 2024. PMID: 38995433
-
Cyclins and CDKs in the regulation of meiosis-specific events.Front Cell Dev Biol. 2022 Nov 29;10:1069064. doi: 10.3389/fcell.2022.1069064. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36523509 Free PMC article. Review.
-
Downregulation of Barley Regulator of Telomere Elongation Helicase 1 Alters the Distribution of Meiotic Crossovers.Front Plant Sci. 2021 Sep 30;12:745070. doi: 10.3389/fpls.2021.745070. eCollection 2021. Front Plant Sci. 2021. PMID: 34659314 Free PMC article.

