Epstein-Barr Virus Nuclear Antigen 1 Recruits Cyclophilin A to Facilitate the Replication of Viral DNA Genome
- PMID: 31921057
- PMCID: PMC6923202
- DOI: 10.3389/fmicb.2019.02879
Epstein-Barr Virus Nuclear Antigen 1 Recruits Cyclophilin A to Facilitate the Replication of Viral DNA Genome
Abstract
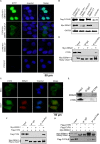
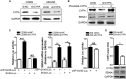
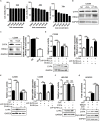
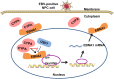
Epstein-Barr virus (EBV) nuclear antigen 1 (EBNA1)-mediated DNA episomal genome replication and persistence are essential for the viral pathogenesis. Cyclophilin A (CYPA) is upregulated in EBV-associated nasopharyngeal carcinoma (NPC) with unknown roles. In the present approach, cytosolic CYPA was found to be bound with EBNA1 into the nucleus. The amino acid 376-459 of the EBNA1 domain was important for the binding. CYPA depletion attenuated and ectopic CYPA expression improved EBNA1 expression in EBV-positive cells. The loss of viral copy number was also accelerated by CYPA consumption in daughter cells during culture passages. Mechanistically, CYPA mediated the connection of EBNA1 with oriP (origin of EBV DNA replication) and subsequent oriP transcription, which is a key step for the initiation of EBV genome replication. Moreover, CYPA overexpression markedly antagonized the connection of EBNA1 to Ubiquitin-specific protease 7 (USP7), which is a strong host barrier with a role of inhibiting EBV genome replication. The PPIase activity of CYPA was required for the promotion of oriP transcription and antagonism with USP7. The results revealed a strategy that EBV recruited a host factor to counteract the host defense, thus facilitating its own latent genome replication. This study provides a new insight into EBV pathogenesis and potential virus-targeted therapeutics in EBV-associated NPC, in which CYPA is upregulated at all stages.
Keywords: Epstein-Barr virus nuclear antigen 1; cyclophilin A; latent genome; pathogenesis; persistence; replication.
Copyright © 2019 Xin, Du, Liu, Xie, Zuo, Yang, Hu, Yue, Zhang, Cao, Zhu and Lu.
Figures



Similar articles
-
B Cell-Specific Transcription Activator PAX5 Recruits p300 To Support EBNA1-Driven Transcription.J Virol. 2020 Mar 17;94(7):e02028-19. doi: 10.1128/JVI.02028-19. Print 2020 Mar 17. J Virol. 2020. PMID: 31941781 Free PMC article.
-
Replication licensing of the EBV oriP minichromosome.Curr Top Microbiol Immunol. 2001;258:13-33. doi: 10.1007/978-3-642-56515-1_2. Curr Top Microbiol Immunol. 2001. PMID: 11443858 Review.
-
EBNA1-mediated recruitment of a histone H2B deubiquitylating complex to the Epstein-Barr virus latent origin of DNA replication.PLoS Pathog. 2009 Oct;5(10):e1000624. doi: 10.1371/journal.ppat.1000624. Epub 2009 Oct 16. PLoS Pathog. 2009. PMID: 19834552 Free PMC article.
-
Carcinoma-risk variant of EBNA1 deregulates Epstein-Barr Virus episomal latency.Oncotarget. 2017 Jan 31;8(5):7248-7264. doi: 10.18632/oncotarget.14540. Oncotarget. 2017. PMID: 28077791 Free PMC article.
-
[Epstein-Barr Virus Genome Replication as a Molecular Target for Cancer Therapy].Yakugaku Zasshi. 2019;139(1):63-67. doi: 10.1248/yakushi.18-00164-1. Yakugaku Zasshi. 2019. PMID: 30606931 Review. Japanese.
Cited by
-
Epstein-Barr Virus Synergizes with BRD7 to Conquer c-Myc-Mediated Viral Latency Maintenance via Chromatin Remodeling.Microbiol Spectr. 2023 Feb 2;11(2):e0123722. doi: 10.1128/spectrum.01237-22. Online ahead of print. Microbiol Spectr. 2023. PMID: 36728436 Free PMC article.
-
Cyclophilin A binds to AKT1 and facilitates the tumorigenicity of Epstein-Barr virus by mediating the activation of AKT/mTOR/NF-κB positive feedback loop.Virol Sin. 2022 Dec;37(6):913-921. doi: 10.1016/j.virs.2022.09.001. Epub 2022 Sep 6. Virol Sin. 2022. PMID: 36075565 Free PMC article.
-
Molecular Mechanisms of DUBs Regulation in Signaling and Disease.Int J Mol Sci. 2021 Jan 20;22(3):986. doi: 10.3390/ijms22030986. Int J Mol Sci. 2021. PMID: 33498168 Free PMC article. Review.
-
Cellular Deubiquitylating Enzyme: A Regulatory Factor of Antiviral Innate Immunity.Front Microbiol. 2021 Dec 13;12:805223. doi: 10.3389/fmicb.2021.805223. eCollection 2021. Front Microbiol. 2021. PMID: 34966378 Free PMC article. Review.
-
Cyclophilin A Inhibits Human Respiratory Syncytial Virus (RSV) Replication by Binding to RSV-N through Its PPIase Activity.J Virol. 2021 Jul 12;95(15):e0056321. doi: 10.1128/JVI.00563-21. Epub 2021 Jul 12. J Virol. 2021. PMID: 34011546 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials

