The core clock gene, Bmal1, and its downstream target, the SNARE regulatory protein secretagogin, are necessary for circadian secretion of glucagon-like peptide-1
- PMID: 31918914
- PMCID: PMC6920326
- DOI: 10.1016/j.molmet.2019.11.004
The core clock gene, Bmal1, and its downstream target, the SNARE regulatory protein secretagogin, are necessary for circadian secretion of glucagon-like peptide-1
Abstract
Objectives: The incretin hormone glucagon-like peptide-1 (GLP-1) is secreted from intestinal L-cells upon nutrient intake. While recent evidence has shown that GLP-1 is released in a circadian manner in rats, whether this occurs in mice and if this pattern is regulated by the circadian clock remain to be elucidated. Furthermore, although circadian GLP-1 secretion parallels expression of the core clock gene Bmal1, the link between the two remains largely unknown. Secretagogin (Scgn) is an exocytotic SNARE regulatory protein that demonstrates circadian expression and is essential for insulin secretion from β-cells. The objective of the current study was to establish the necessity of the core clock gene Bmal1 and the SNARE protein SCGN as essential regulators of circadian GLP-1 secretion.
Methods: Oral glucose tolerance tests were conducted at different times of the day on 4-hour fasted C57BL/6J, Bmal1 wild-type, and Bmal1 knockout mice. Mass spectrometry, RNA-seq, qRT-PCR and/or microarray analyses, and immunostaining were conducted on murine (m) and human (h) primary L-cells and mGLUTag and hNCI-H716 L-cell lines. At peak and trough GLP-1 secretory time points, the mGLUTag cells were co-stained for SCGN and a membrane-marker, ChIP was used to analyze BMAL1 binding sites in the Scgn promoter, protein interaction with SCGN was tested by co-immunoprecipitation, and siRNA was used to knockdown Scgn for GLP-1 secretion assay.
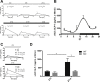
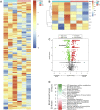
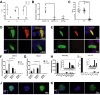
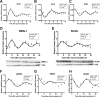
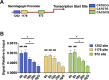
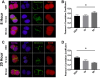
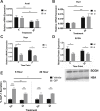
Results: C57BL/6J mice displayed a circadian rhythm in GLP-1 secretion that peaked at the onset of their feeding period. Rhythmic GLP-1 release was impaired in Bmal1 knockout (KO) mice as compared to wild-type controls at the peak (p < 0.05) but not at the trough secretory time point. Microarray identified SNARE and transport vesicle pathways as highly upregulated in mGLUTag L-cells at the peak time point of GLP-1 secretion (p < 0.001). Mass spectrometry revealed that SCGN was also increased at this time (p < 0.001), while RNA-seq, qRT-PCR, and immunostaining demonstrated Scgn expression in all human and murine primary L-cells and cell lines. The mGLUTag and hNCI-H716 L-cells exhibited circadian rhythms in Scgn expression (p < 0.001). The ChIP analysis demonstrated increased binding of BMAL1 only at the peak of Scgn expression (p < 0.01). Immunocytochemistry showed the translocation of SCGN to the cell membrane after stimulation at the peak time point only (p < 0.05), while CoIP showed that SCGN was pulled down with SNAP25 and β-actin, but only the latter interaction was time-dependent (p < 0.05). Finally, Scgn siRNA-treated cells demonstrated significantly blunted GLP-1 secretion (p < 0.01) in response to stimulation at the peak time point only.
Conclusions: These data demonstrate, for the first time, that mice display a circadian pattern in GLP-1 secretion, which is impaired in Bmal1 knockout mice, and that Bmal1 regulation of Scgn expression plays an essential role in the circadian release of the incretin hormone GLP-1.
Keywords: Bmal1; Circadian; GLP-1; L-cell; Secretagogin; Secretion.
Copyright © 2019 The Author(s). Published by Elsevier GmbH.. All rights reserved.
Figures

Similar articles
-
The Cytoskeletal Transport Protein, Secretagogin, Is Essential for Diurnal Glucagon-like Peptide-1 Secretion in Mice.Endocrinology. 2022 Oct 11;163(11):bqac142. doi: 10.1210/endocr/bqac142. Endocrinology. 2022. PMID: 36036556
-
L-cell Arntl is required for rhythmic glucagon-like peptide-1 secretion and maintenance of intestinal homeostasis.Mol Metab. 2021 Dec;54:101340. doi: 10.1016/j.molmet.2021.101340. Epub 2021 Sep 11. Mol Metab. 2021. PMID: 34520858 Free PMC article.
-
Essential Role of Syntaxin-Binding Protein-1 in the Regulation of Glucagon-Like Peptide-1 Secretion.Endocrinology. 2020 May 1;161(5):bqaa039. doi: 10.1210/endocr/bqaa039. Endocrinology. 2020. PMID: 32141504 Free PMC article.
-
Glucagon-like peptide-1: The missing link in the metabolic clock?J Diabetes Investig. 2016 Apr;7 Suppl 1(Suppl 1):70-5. doi: 10.1111/jdi.12477. Epub 2016 Mar 14. J Diabetes Investig. 2016. PMID: 27186359 Free PMC article. Review.
-
Effects of BMAL1 Manipulation on the Brain's Master Circadian Clock and Behavior.Yale J Biol Med. 2019 Jun 27;92(2):251-258. eCollection 2019 Jun. Yale J Biol Med. 2019. PMID: 31249486 Free PMC article. Review.
Cited by
-
Disrupted and Elevated Circadian Secretion of Glucagon-Like Peptide-1 in a Murine Model of Type 2 Diabetes.Endocrinology. 2022 Sep 1;163(9):bqac118. doi: 10.1210/endocr/bqac118. Endocrinology. 2022. PMID: 35876276 Free PMC article.
-
Timing Matters: The Interplay between Early Mealtime, Circadian Rhythms, Gene Expression, Circadian Hormones, and Metabolism-A Narrative Review.Clocks Sleep. 2023 Sep 6;5(3):507-535. doi: 10.3390/clockssleep5030034. Clocks Sleep. 2023. PMID: 37754352 Free PMC article. Review.
-
Role of High Energy Breakfast "Big Breakfast Diet" in Clock Gene Regulation of Postprandial Hyperglycemia and Weight Loss in Type 2 Diabetes.Nutrients. 2021 May 5;13(5):1558. doi: 10.3390/nu13051558. Nutrients. 2021. PMID: 34063109 Free PMC article. Review.
-
Advancements in research on the association between the biological CLOCK and type 2 diabetes.Front Endocrinol (Lausanne). 2024 May 30;15:1320605. doi: 10.3389/fendo.2024.1320605. eCollection 2024. Front Endocrinol (Lausanne). 2024. PMID: 38872971 Free PMC article. Review.
-
Circadian hormone secretion of enteroendocrine cells: implication on pregnancy status.Front Endocrinol (Lausanne). 2023 May 10;14:1106382. doi: 10.3389/fendo.2023.1106382. eCollection 2023. Front Endocrinol (Lausanne). 2023. PMID: 37234809 Free PMC article. Review.
References
-
- Bass J., Lazar M.A. Circadian time signatures of fitness and Disease. Science (New York, N.Y.) 2016;354(6315):994–999. - PubMed
-
- Tahara Y., Shibata S. Circadian rhythms of liver physiology and Disease: experimental and clinical evidence. Nature Reviews Gastroenterology and Hepatology. 2016;13(4):217–226. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

