Using Phaser and ensembles to improve the performance of SIMBAD
- PMID: 31909738
- PMCID: PMC6939438
- DOI: 10.1107/S2059798319015031
Using Phaser and ensembles to improve the performance of SIMBAD
Abstract
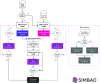
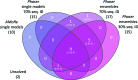
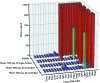
The conventional approach to search-model identification in molecular replacement (MR) is to screen a database of known structures using the target sequence. However, this strategy is not always effective, for example when the relationship between sequence and structural similarity fails or when the crystal contents are not those expected. An alternative approach is to identify suitable search models directly from the experimental data. SIMBAD is a sequence-independent MR pipeline that uses either a crystal lattice search or MR functions to directly locate suitable search models from databases. The previous version of SIMBAD used the fast AMoRe rotation-function search. Here, a new version of SIMBAD which makes use of Phaser and its likelihood scoring to improve the sensitivity of the pipeline is presented. It is shown that the additional compute time potentially required by the more sophisticated scoring is counterbalanced by the greater sensitivity, allowing more cases to trigger early-termination criteria, rather than running to completion. Using Phaser solved 17 out of 25 test cases in comparison to the ten solved with AMoRe, and it is shown that use of ensemble search models produces additional performance benefits.
Keywords: SIMBAD; contaminants; ensembles; molecular-replacement pipeline; sequence independent; structure solution.
open access.
Figures

Similar articles
-
Molecular replacement using structure predictions from databases.Acta Crystallogr D Struct Biol. 2019 Dec 1;75(Pt 12):1051-1062. doi: 10.1107/S2059798319013962. Epub 2019 Nov 19. Acta Crystallogr D Struct Biol. 2019. PMID: 31793899 Free PMC article.
-
SIMBAD: a sequence-independent molecular-replacement pipeline.Acta Crystallogr D Struct Biol. 2018 Jul 1;74(Pt 7):595-605. doi: 10.1107/S2059798318005752. Epub 2018 Jun 8. Acta Crystallogr D Struct Biol. 2018. PMID: 29968670 Free PMC article.
-
Ensembles generated from crystal structures of single distant homologues solve challenging molecular-replacement cases in AMPLE.Acta Crystallogr D Struct Biol. 2018 Mar 1;74(Pt 3):183-193. doi: 10.1107/S2059798318002310. Epub 2018 Mar 2. Acta Crystallogr D Struct Biol. 2018. PMID: 29533226 Free PMC article.
-
Phaser.MRage: automated molecular replacement.Acta Crystallogr D Biol Crystallogr. 2013 Nov;69(Pt 11):2276-86. doi: 10.1107/S0907444913022750. Epub 2013 Oct 18. Acta Crystallogr D Biol Crystallogr. 2013. PMID: 24189240 Free PMC article. Review.
-
Acknowledging Errors: Advanced Molecular Replacement with Phaser.Methods Mol Biol. 2017;1607:421-453. doi: 10.1007/978-1-4939-7000-1_18. Methods Mol Biol. 2017. PMID: 28573584 Review.
Cited by
-
Molecular replacement using structure predictions from databases.Acta Crystallogr D Struct Biol. 2019 Dec 1;75(Pt 12):1051-1062. doi: 10.1107/S2059798319013962. Epub 2019 Nov 19. Acta Crystallogr D Struct Biol. 2019. PMID: 31793899 Free PMC article.
-
Recent developments in MrBUMP: better search-model preparation, graphical interaction with search models, and solution improvement and assessment.Acta Crystallogr D Struct Biol. 2018 Mar 1;74(Pt 3):167-182. doi: 10.1107/S2059798318003455. Epub 2018 Mar 6. Acta Crystallogr D Struct Biol. 2018. PMID: 29533225 Free PMC article.
-
Introduction to molecular replacement: a time perspective.Acta Crystallogr D Struct Biol. 2021 Jul 1;77(Pt 7):867-879. doi: 10.1107/S2059798321004368. Epub 2021 Jun 18. Acta Crystallogr D Struct Biol. 2021. PMID: 34196614 Free PMC article. Review.
-
findMySequence: a neural-network-based approach for identification of unknown proteins in X-ray crystallography and cryo-EM.IUCrJ. 2021 Dec 1;9(Pt 1):86-97. doi: 10.1107/S2052252521011088. eCollection 2022 Jan 1. IUCrJ. 2021. PMID: 35059213 Free PMC article.
-
Identification, structure determination and analysis of Mycobacterium smegmatis acyl-carrier protein synthase (AcpS) crystallized serendipitously.Acta Crystallogr F Struct Biol Commun. 2022 Jul 1;78(Pt 7):252-264. doi: 10.1107/S2053230X22005738. Epub 2022 Jun 15. Acta Crystallogr F Struct Biol Commun. 2022. PMID: 35787552 Free PMC article.
References
-
- Adams, P. D., Afonine, P. V., Bunkóczi, G., Chen, V. B., Davis, I. W., Echols, N., Headd, J. J., Hung, L.-W., Kapral, G. J., Grosse-Kunstleve, R. W., McCoy, A. J., Moriarty, N. W., Oeffner, R., Read, R. J., Richardson, D. C., Richardson, J. S., Terwilliger, T. C. & Zwart, P. H. (2010). Acta Cryst. D66, 213–221. - PMC - PubMed
-
- Bibby, J., Keegan, R. M., Mayans, O., Winn, M. D. & Rigden, D. J. (2012). Acta Cryst. D68, 1622–1631. - PubMed
-
- Chen, Y. W., Dodson, E. J. & Kleywegt, G. J. (2000). Structure, 8, R213–R220. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

