Identification and characterization of extrachromosomal circular DNA in maternal plasma
- PMID: 31900366
- PMCID: PMC6983429
- DOI: 10.1073/pnas.1914949117
Identification and characterization of extrachromosomal circular DNA in maternal plasma
Abstract
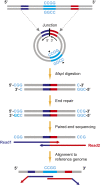
We explored the presence of extrachromosomal circular DNA (eccDNA) in the plasma of pregnant women. Through sequencing following either restriction enzyme or Tn5 transposase treatment, we identified eccDNA molecules in the plasma of pregnant women. These eccDNA molecules showed bimodal size distributions peaking at ∼202 and ∼338 bp with distinct 10-bp periodicity observed throughout the size ranges within both peaks, suggestive of their nucleosomal origin. Also, the predominance of the 338-bp peak of eccDNA indicated that eccDNA had a larger size distribution than linear DNA in human plasma. Moreover, eccDNA of fetal origin were shorter than the maternal eccDNA. Genomic annotation of the overall population of eccDNA molecules revealed a preference of these molecules to be generated from 5'-untranslated regions (5'-UTRs), exonic regions, and CpG island regions. Two sets of trinucleotide repeat motifs flanking the junctional sites of eccDNA supported multiple possible models for eccDNA generation. This work highlights the topologic analysis of plasma DNA, which is an emerging direction for circulating nucleic acid research and applications.
Keywords: cell-free DNA; eccDNA; noninvasive prenatal testing; plasma DNA topologics.
Copyright © 2020 the Author(s). Published by PNAS.
Conflict of interest statement
Competing interest statement: S.T.K.S., P.J., J.D., L.J., K.C.A.C., R.W.K.C. and Y.M.D.L. have filed a patent application based on the data generated from this work.
Figures

Similar articles
-
Characteristics of Fetal Extrachromosomal Circular DNA in Maternal Plasma: Methylation Status and Clearance.Clin Chem. 2021 Apr 29;67(5):788-796. doi: 10.1093/clinchem/hvaa326. Clin Chem. 2021. PMID: 33615350
-
Molecular characterization of cell-free eccDNAs in human plasma.Sci Rep. 2017 Sep 8;7(1):10968. doi: 10.1038/s41598-017-11368-w. Sci Rep. 2017. PMID: 28887493 Free PMC article.
-
Effects of nucleases on cell-free extrachromosomal circular DNA.JCI Insight. 2022 Apr 22;7(8):e156070. doi: 10.1172/jci.insight.156070. JCI Insight. 2022. PMID: 35451374 Free PMC article.
-
Extrachromosomal circular DNA in eukaryotes: possible involvement in the plasticity of tandem repeats.Cytogenet Genome Res. 2009;124(3-4):327-38. doi: 10.1159/000218136. Epub 2009 Jun 25. Cytogenet Genome Res. 2009. PMID: 19556784 Review.
-
Extrachromosomal circular DNA: biogenesis, structure, functions and diseases.Signal Transduct Target Ther. 2022 Oct 2;7(1):342. doi: 10.1038/s41392-022-01176-8. Signal Transduct Target Ther. 2022. PMID: 36184613 Free PMC article. Review.
Cited by
-
Bioinformatics advances in eccDNA identification and analysis.Oncogene. 2024 Oct;43(41):3021-3036. doi: 10.1038/s41388-024-03138-6. Epub 2024 Aug 29. Oncogene. 2024. PMID: 39209966 Review.
-
Predicting gestational diabetes mellitus risk at 11-13 weeks' gestation: the role of extrachromosomal circular DNA.Cardiovasc Diabetol. 2024 Aug 7;23(1):289. doi: 10.1186/s12933-024-02381-1. Cardiovasc Diabetol. 2024. PMID: 39113025 Free PMC article.
-
Characterization of extrachromosomal circular DNAs in plasma of patients with clear cell renal cell carcinoma.World J Urol. 2024 May 16;42(1):328. doi: 10.1007/s00345-024-05031-z. World J Urol. 2024. PMID: 38753087
-
Extrachromosomal Circular DNA: An Emerging Potential Biomarker for Inflammatory Bowel Diseases?Genes (Basel). 2024 Mar 26;15(4):414. doi: 10.3390/genes15040414. Genes (Basel). 2024. PMID: 38674347 Free PMC article. Review.
-
Categorizing Extrachromosomal Circular DNA as Biomarkers in Serum of Cancer.Biomolecules. 2024 Apr 17;14(4):488. doi: 10.3390/biom14040488. Biomolecules. 2024. PMID: 38672504 Free PMC article. Review.
References
-
- Lo Y. M. D., et al. , Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci. Transl. Med. 2, 61ra91 (2010). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

