The five dimensions of receptor pharmacology exemplified by melatonin receptors: An opinion
- PMID: 31893125
- PMCID: PMC6935684
- DOI: 10.1002/prp2.556
The five dimensions of receptor pharmacology exemplified by melatonin receptors: An opinion
Abstract
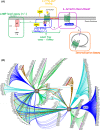
Receptology has been complicated with enhancements in our knowledge of G-protein-coupled-receptor (GPCR) biochemistry. This complexity is exemplified by the pharmacology of melatonin receptors. Here, we describe the complexity of GPCR biochemistry in five dimensions: (a) receptor expression, particularly in organs/tissues that are only partially understood; (b) ligands and receptor-associated proteins (interactome); (c) receptor function, which might be more complex than the known G-protein-coupled systems; (d) ligand bias, which favors a particular pathway; and (e) receptor dimerization, which might concern all receptors coexpressed in the same cell. Thus, receptor signaling might be modified or modulated, depending on the nature of the receptor complex. Fundamental studies are needed to clarify these points and find new ways to tackle receptor functionality. This opinion article emphasizes the global questions attached to new descriptions of GPCRs and aims to raise our awareness of the tremendous complexity of modern receptology.
Keywords: GPCRs; antagonism; biased ligands; dimerization; expression; interactome; melatonin; review.
© 2019 The Authors. Pharmacology Research & Perspectives published by John Wiley & Sons Ltd, British Pharmacological Society and American Society for Pharmacology and Experimental Therapeutics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Melatonin receptors: molecular pharmacology and signalling in the context of system bias.Br J Pharmacol. 2018 Aug;175(16):3263-3280. doi: 10.1111/bph.13950. Epub 2017 Aug 17. Br J Pharmacol. 2018. PMID: 28707298 Free PMC article. Review.
-
Melatonin Receptor Signaling: Impact of Receptor Oligomerization on Receptor Function.Int Rev Cell Mol Biol. 2018;338:59-77. doi: 10.1016/bs.ircmb.2018.02.002. Epub 2018 Apr 5. Int Rev Cell Mol Biol. 2018. PMID: 29699692 Review.
-
Melatonin receptors, heterodimerization, signal transduction and binding sites: what's new?Br J Pharmacol. 2008 Jul;154(6):1182-95. doi: 10.1038/bjp.2008.184. Epub 2008 May 19. Br J Pharmacol. 2008. PMID: 18493248 Free PMC article. Review.
-
Are G protein-coupled receptor heterodimers of physiological relevance?--Focus on melatonin receptors.Chronobiol Int. 2006;23(1-2):419-26. doi: 10.1080/07420520500521863. Chronobiol Int. 2006. PMID: 16687315 Review.
-
Two neuroendocrine G protein-coupled receptor molecules, somatostatin and melatonin: Physiology of signal transduction and therapeutic perspectives.J Cell Physiol. 2021 Apr;236(4):2505-2518. doi: 10.1002/jcp.30062. Epub 2020 Sep 28. J Cell Physiol. 2021. PMID: 32989768 Review.
Cited by
-
Functionality of Melatonin Receptors: Internalization.Methods Mol Biol. 2022;2550:189-193. doi: 10.1007/978-1-0716-2593-4_23. Methods Mol Biol. 2022. PMID: 36180692
-
MT1 Receptor Signaling Pathways by Impedance Measurement.Methods Mol Biol. 2022;2550:201-206. doi: 10.1007/978-1-0716-2593-4_25. Methods Mol Biol. 2022. PMID: 36180694
-
N-(Anilinoethyl)amide Melatonergic Ligands with Improved Water Solubility and Metabolic Stability.ChemMedChem. 2021 Oct 6;16(19):3071-3082. doi: 10.1002/cmdc.202100405. Epub 2021 Jul 26. ChemMedChem. 2021. PMID: 34213063 Free PMC article.
-
Evaluation of neurobiological and antioxidant effects of novel melatonin analogs in mice.Saudi Pharm J. 2020 Dec;28(12):1566-1579. doi: 10.1016/j.jsps.2020.10.004. Epub 2020 Oct 21. Saudi Pharm J. 2020. PMID: 33424250 Free PMC article.
-
MCH-R1 Antagonist GPS18169, a Pseudopeptide, Is a Peripheral Anti-Obesity Agent in Mice.Molecules. 2021 Feb 27;26(5):1291. doi: 10.3390/molecules26051291. Molecules. 2021. PMID: 33673598 Free PMC article.
References
-
- Zhou XE, Melcher K, Xu HE. Understanding the GPCR biased signaling through G protein and arrestin complex structures. Curr Opin Struct Biol. 2017;45:150‐159. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

