Regulatory Role of PlaR (YiaJ) for Plant Utilization in Escherichia coli K-12
- PMID: 31892694
- PMCID: PMC6958661
- DOI: 10.1038/s41598-019-56886-x
Regulatory Role of PlaR (YiaJ) for Plant Utilization in Escherichia coli K-12
Erratum in
-
Author Correction: Regulatory Role of PlaR (YiaJ) for Plant Utilization in Escherichia coli K-12.Sci Rep. 2020 Feb 14;10(1):2997. doi: 10.1038/s41598-020-59905-4. Sci Rep. 2020. PMID: 32060397 Free PMC article.
Abstract
Outside a warm-blooded animal host, the enterobacterium Escherichia coli K-12 is also able to grow and survive in stressful nature. The major organic substance in nature is plant, but the genetic system of E. coli how to utilize plant-derived materials as nutrients is poorly understood. Here we describe the set of regulatory targets for uncharacterized IclR-family transcription factor YiaJ on the E. coli genome, using gSELEX screening system. Among a total of 18 high-affinity binding targets of YiaJ, the major regulatory target was identified to be the yiaLMNOPQRS operon for utilization of ascorbate from fruits and galacturonate from plant pectin. The targets of YiaJ also include the genes involved in the utilization for other plant-derived materials as nutrients such as fructose, sorbitol, glycerol and fructoselysine. Detailed in vitro and in vivo analyses suggest that L-ascorbate and α-D-galacturonate are the effector ligands for regulation of YiaJ function. These findings altogether indicate that YiaJ plays a major regulatory role in expression of a set of the genes for the utilization of plant-derived materials as nutrients for survival. PlaR was also suggested to play protecting roles of E. coli under stressful environments in nature, including the formation of biofilm. We then propose renaming YiaJ to PlaR (regulator of plant utilization). The natural hosts of enterobacterium Escherichia coli are warm-blooded animals, but even outside hosts, E. coli can survive even under stressful environments. On earth, the most common organic materials to be used as nutrients by E. coli are plant-derived components, but up to the present time, the genetic system of E. coli for plant utilization is poorly understand. In the course of gSELEX screening of the regulatory targets for hitherto uncharacterized TFs, we identified in this study the involvement of the IclR-family YiaJ in the regulation of about 20 genes or operons, of which the majority are related to the catabolism of plant-derived materials such as ascorbate, galacturonate, sorbitol, fructose and fructoselysine. Therefore, we propose to rename YiaJ to PlaR (regulator of plant utilization).
Conflict of interest statement
The authors declare no competing interests.
Figures
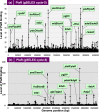
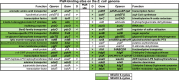



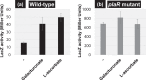
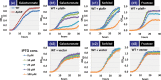

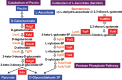
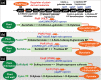
Similar articles
-
Transcription Factor SrsR (YgfI) Is a Novel Regulator for the Stress-Response Genes in Stationary Phase in Escherichia coli K-12.Int J Mol Sci. 2022 May 27;23(11):6055. doi: 10.3390/ijms23116055. Int J Mol Sci. 2022. PMID: 35682733 Free PMC article.
-
Novel regulators of the csgD gene encoding the master regulator of biofilm formation in Escherichia coli K-12.Microbiology (Reading). 2020 Sep;166(9):880-890. doi: 10.1099/mic.0.000947. Microbiology (Reading). 2020. PMID: 32649279
-
Systematic discovery of uncharacterized transcription factors in Escherichia coli K-12 MG1655.Nucleic Acids Res. 2018 Nov 16;46(20):10682-10696. doi: 10.1093/nar/gky752. Nucleic Acids Res. 2018. PMID: 30137486 Free PMC article.
-
Hierarchy of transcription factor network in Escherichia coli K-12: H-NS-mediated silencing and Anti-silencing by global regulators.FEMS Microbiol Rev. 2021 Nov 23;45(6):fuab032. doi: 10.1093/femsre/fuab032. FEMS Microbiol Rev. 2021. PMID: 34196371 Review.
-
Contribution of rpoS and bolA genes in biofilm formation in Escherichia coli K-12 MG1655.Mol Cell Biochem. 2010 Sep;342(1-2):207-13. doi: 10.1007/s11010-010-0485-7. Epub 2010 May 18. Mol Cell Biochem. 2010. PMID: 20480211 Review.
Cited by
-
Transcription Factor SrsR (YgfI) Is a Novel Regulator for the Stress-Response Genes in Stationary Phase in Escherichia coli K-12.Int J Mol Sci. 2022 May 27;23(11):6055. doi: 10.3390/ijms23116055. Int J Mol Sci. 2022. PMID: 35682733 Free PMC article.
-
Escherichia coli transcription factors of unknown function: sequence features and possible evolutionary relationships.PeerJ. 2022 Jul 20;10:e13772. doi: 10.7717/peerj.13772. eCollection 2022. PeerJ. 2022. PMID: 35880217 Free PMC article.
-
Persistence and plasticity in bacterial gene regulation.Nat Methods. 2021 Dec;18(12):1499-1505. doi: 10.1038/s41592-021-01312-2. Epub 2021 Nov 25. Nat Methods. 2021. PMID: 34824476
-
Expanded roles of lactate-sensing LldR in transcription regulation of the Escherichia coli K-12 genome: lactate utilisation and acid resistance.Microb Genom. 2023 May;9(5):mgen001015. doi: 10.1099/mgen.0.001015. Microb Genom. 2023. PMID: 37219924 Free PMC article.
-
Expanded roles of pyruvate-sensing PdhR in transcription regulation of the Escherichia coli K-12 genome: fatty acid catabolism and cell motility.Microb Genom. 2020 Oct;6(10):mgen000442. doi: 10.1099/mgen.0.000442. Microb Genom. 2020. PMID: 32975502 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

