Loss of the disease-associated glycosyltransferase Galnt3 alters Muc10 glycosylation and the composition of the oral microbiome
- PMID: 31882545
- PMCID: PMC6996895
- DOI: 10.1074/jbc.RA119.009807
Loss of the disease-associated glycosyltransferase Galnt3 alters Muc10 glycosylation and the composition of the oral microbiome
Abstract
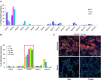
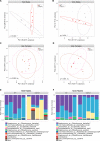
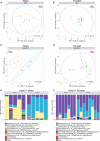
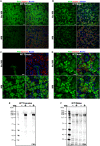
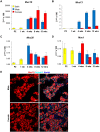
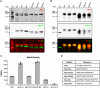
The importance of the microbiome in health and its disruption in disease is continuing to be elucidated. However, the multitude of host and environmental factors that influence the microbiome are still largely unknown. Here, we examined UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase 3 (Galnt3)-deficient mice, which serve as a model for the disease hyperphosphatemic familial tumoral calcinosis (HFTC). In HFTC, loss of GALNT3 activity in the bone is thought to lead to altered glycosylation of the phosphate-regulating hormone fibroblast growth factor 23 (FGF23), resulting in hyperphosphatemia and subdermal calcified tumors. However, GALNT3 is expressed in other tissues in addition to bone, suggesting that systemic loss could result in other pathologies. Using semiquantitative real-time PCR, we found that Galnt3 is the major O-glycosyltransferase expressed in the secretory cells of salivary glands. Additionally, 16S rRNA gene sequencing revealed that the loss of Galnt3 resulted in changes in the structure, composition, and stability of the oral microbiome. Moreover, we identified the major secreted salivary mucin, Muc10, as an in vivo substrate of Galnt3. Given that mucins and their O-glycans are known to interact with various microbes, our results suggest that loss of Galnt3 decreases glycosylation of Muc10, which alters the composition and stability of the oral microbiome. Considering that oral findings have been documented in HFTC patients, our study suggests that investigating GALNT3-mediated changes in the oral microbiome may be warranted.
Keywords: Galnt3; Muc10; O-glycosylation; bone; glycan; glycosylation; glycosyltransferase; hyperphosphatemic familial tumoral calcinosis (HFTC); microbiome; mucin; post-translational modification; saliva; salivary protein; submandibular gland.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures

Similar articles
-
Ablation of the Galnt3 gene leads to low-circulating intact fibroblast growth factor 23 (Fgf23) concentrations and hyperphosphatemia despite increased Fgf23 expression.Endocrinology. 2009 Jun;150(6):2543-50. doi: 10.1210/en.2008-0877. Epub 2009 Feb 12. Endocrinology. 2009. PMID: 19213845 Free PMC article.
-
In vivo mapping of the mouse Galnt3-specific O-glycoproteome.J Biol Chem. 2024 Sep;300(9):107628. doi: 10.1016/j.jbc.2024.107628. Epub 2024 Aug 2. J Biol Chem. 2024. PMID: 39098533 Free PMC article.
-
Long-term clinical outcome and phenotypic variability in hyperphosphatemic familial tumoral calcinosis and hyperphosphatemic hyperostosis syndrome caused by a novel GALNT3 mutation; case report and review of the literature.BMC Genet. 2014 Sep 24;15:98. doi: 10.1186/s12863-014-0098-3. BMC Genet. 2014. PMID: 25249269 Free PMC article. Review.
-
Hyperphosphatemic Familial Tumoral Calcinosis in Two Siblings with a Novel Mutation in GALNT3 Gene: Experience from Southern Turkey.J Clin Res Pediatr Endocrinol. 2019 Feb 20;11(1):94-99. doi: 10.4274/jcrpe.galenos.2018.2018.0134. Epub 2018 Jul 17. J Clin Res Pediatr Endocrinol. 2019. PMID: 30015621 Free PMC article.
-
Hyperphosphatemic familial tumoral calcinosis: genetic models of deficient FGF23 action.Curr Osteoporos Rep. 2015 Apr;13(2):78-87. doi: 10.1007/s11914-015-0254-3. Curr Osteoporos Rep. 2015. PMID: 25656441 Review.
Cited by
-
Mucin-Type O-GalNAc Glycosylation in Health and Disease.Adv Exp Med Biol. 2021;1325:25-60. doi: 10.1007/978-3-030-70115-4_2. Adv Exp Med Biol. 2021. PMID: 34495529
-
Multi-omics analysis reveals the effects of microbiota on oral homeostasis.Front Immunol. 2022 Sep 20;13:1005992. doi: 10.3389/fimmu.2022.1005992. eCollection 2022. Front Immunol. 2022. PMID: 36211346 Free PMC article.
-
Differential splicing of the lectin domain of an O-glycosyltransferase modulates both peptide and glycopeptide preferences.J Biol Chem. 2020 Aug 28;295(35):12525-12536. doi: 10.1074/jbc.RA120.014700. Epub 2020 Jul 15. J Biol Chem. 2020. PMID: 32669364 Free PMC article.
-
Development of a cell-based pathway modulator screening system to screen the targeted cancer therapeutic candidates.Hum Cell. 2021 Mar;34(2):445-456. doi: 10.1007/s13577-020-00476-x. Epub 2021 Jan 6. Hum Cell. 2021. PMID: 33405176
-
Salivary gland function, development, and regeneration.Physiol Rev. 2022 Jul 1;102(3):1495-1552. doi: 10.1152/physrev.00015.2021. Epub 2022 Mar 28. Physiol Rev. 2022. PMID: 35343828 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

