Exploring the changing landscape of cell-to-cell variation after CTCF knockdown via single cell RNA-seq
- PMID: 31878887
- PMCID: PMC6933653
- DOI: 10.1186/s12864-019-6379-5
Exploring the changing landscape of cell-to-cell variation after CTCF knockdown via single cell RNA-seq
Abstract
Background: CCCTC-Binding Factor (CTCF), also known as 11-zinc finger protein, participates in many cellular processes, including insulator activity, transcriptional regulation and organization of chromatin architecture. Based on single cell flow cytometry and single cell RNA-FISH analyses, our previous study showed that deletion of CTCF binding site led to a significantly increase of cellular variation of its target gene. However, the effect of CTCF on genome-wide landscape of cell-to-cell variation remains unclear.
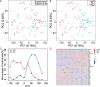
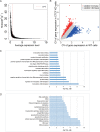
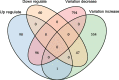
Results: We knocked down CTCF in EL4 cells using shRNA, and conducted single cell RNA-seq on both wild type (WT) cells and CTCF-Knockdown (CTCF-KD) cells using Fluidigm C1 system. Principal component analysis of single cell RNA-seq data showed that WT and CTCF-KD cells concentrated in two different clusters on PC1, indicating that gene expression profiles of WT and CTCF-KD cells were systematically different. Interestingly, GO terms including regulation of transcription, DNA binding, zinc finger and transcription factor binding were significantly enriched in CTCF-KD-specific highly variable genes, implying tissue-specific genes such as transcription factors were highly sensitive to CTCF level. The dysregulation of transcription factors potentially explains why knockdown of CTCF leads to systematic change of gene expression. In contrast, housekeeping genes such as rRNA processing, DNA repair and tRNA processing were significantly enriched in WT-specific highly variable genes, potentially due to a higher cellular variation of cell activity in WT cells compared to CTCF-KD cells. We further found that cellular variation-increased genes were significantly enriched in down-regulated genes, indicating CTCF knockdown simultaneously reduced the expression levels and increased the expression noise of its regulated genes.
Conclusions: To our knowledge, this is the first attempt to explore genome-wide landscape of cellular variation after CTCF knockdown. Our study not only advances our understanding of CTCF function in maintaining gene expression and reducing expression noise, but also provides a framework for examining gene function.
Keywords: CTCF; CTCF knockdown; Cell-to-cell variation; Change of cellular variation; Single cell RNA-seq.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
CTCF Expression is Essential for Somatic Cell Viability and Protection Against Cancer.Int J Mol Sci. 2018 Nov 30;19(12):3832. doi: 10.3390/ijms19123832. Int J Mol Sci. 2018. PMID: 30513694 Free PMC article.
-
Liver-Specific Deletion of Mouse CTCF Leads to Hepatic Steatosis via Augmented PPARγ Signaling.Cell Mol Gastroenterol Hepatol. 2021;12(5):1761-1787. doi: 10.1016/j.jcmgh.2021.07.016. Epub 2021 Aug 4. Cell Mol Gastroenterol Hepatol. 2021. PMID: 34358714 Free PMC article.
-
Acute depletion of CTCF rewires genome-wide chromatin accessibility.Genome Biol. 2021 Aug 24;22(1):244. doi: 10.1186/s13059-021-02466-0. Genome Biol. 2021. PMID: 34429148 Free PMC article.
-
CTCF: a Swiss-army knife for genome organization and transcription regulation.Essays Biochem. 2019 Apr 23;63(1):157-165. doi: 10.1042/EBC20180069. Print 2019 Apr 23. Essays Biochem. 2019. PMID: 30940740 Review.
-
A tour of 3D genome with a focus on CTCF.Semin Cell Dev Biol. 2019 Jun;90:4-11. doi: 10.1016/j.semcdb.2018.07.020. Epub 2018 Jul 23. Semin Cell Dev Biol. 2019. PMID: 30031214 Review.
Cited by
-
Comprehensive mapping of alternative polyadenylation site usage and its dynamics at single-cell resolution.Proc Natl Acad Sci U S A. 2022 Dec 6;119(49):e2113504119. doi: 10.1073/pnas.2113504119. Epub 2022 Dec 1. Proc Natl Acad Sci U S A. 2022. PMID: 36454750 Free PMC article.
-
Integrated decoding hematopoiesis and leukemogenesis using single-cell sequencing and its medical implication.Cell Discov. 2021 Jan 5;7(1):2. doi: 10.1038/s41421-020-00223-4. Cell Discov. 2021. PMID: 33408321 Free PMC article.
-
Contributions of transcriptional noise to leukaemia evolution: KAT2A as a case-study.Philos Trans R Soc Lond B Biol Sci. 2024 Apr 22;379(1900):20230052. doi: 10.1098/rstb.2023.0052. Epub 2024 Mar 4. Philos Trans R Soc Lond B Biol Sci. 2024. PMID: 38432321 Free PMC article. Review.
-
Diploid genome architecture revealed by multi-omic data of hybrid mice.Genome Res. 2020 Aug;30(8):1097-1106. doi: 10.1101/gr.257568.119. Epub 2020 Aug 5. Genome Res. 2020. PMID: 32759226 Free PMC article.
-
Loop stacking organizes genome folding from TADs to chromosomes.Mol Cell. 2023 May 4;83(9):1377-1392.e6. doi: 10.1016/j.molcel.2023.04.008. Mol Cell. 2023. PMID: 37146570 Free PMC article.
References
-
- Lobanenkov VV, Nicolas RH, Adler VV, Paterson H, Klenova EM, Polotskaja AV, Goodwin GH. A novel sequence-specific DNA binding protein which interacts with three regularly spaced direct repeats of the CCCTC-motif in the 5′-flanking sequence of the chicken c-myc gene. Oncogene. 1990;5(12):1743–1753. - PubMed
-
- Klenova EM, Nicolas RH, Paterson HF, Carne AF, Heath CM, Goodwin GH, Neiman PE, Lobanenkov VV. CTCF, a conserved nuclear factor required for optimal transcriptional activity of the chicken c-myc gene, is an 11-Zn-finger protein differentially expressed in multiple forms. Mol Cell Biol. 1993;13(12):7612–7624. doi: 10.1128/MCB.13.12.7612. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

