The energetics of protein-lipid interactions as viewed by molecular simulations
- PMID: 31872229
- PMCID: PMC7054751
- DOI: 10.1042/BST20190149
The energetics of protein-lipid interactions as viewed by molecular simulations
Abstract
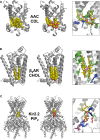
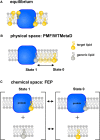
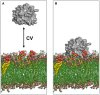
Membranes are formed from a bilayer containing diverse lipid species with which membrane proteins interact. Integral, membrane proteins are embedded in this bilayer, where they interact with lipids from their surroundings, whilst peripheral membrane proteins bind to lipids at the surface of membranes. Lipid interactions can influence the function of membrane proteins, either directly or allosterically. Both experimental (structural) and computational approaches can reveal lipid binding sites on membrane proteins. It is, therefore, important to understand the free energies of these interactions. This affords a more complete view of the engagement of a particular protein with the biological membrane surrounding it. Here, we describe many computational approaches currently in use for this purpose, including recent advances using both free energy and unbiased simulation methods. In particular, we focus on interactions of integral membrane proteins with cholesterol, and with anionic lipids such as phosphatidylinositol 4,5-bis-phosphate and cardiolipin. Peripheral membrane proteins are exemplified via interactions of PH domains with phosphoinositide-containing membranes. We summarise the current state of the field and provide an outlook on likely future directions of investigation.
Keywords: free energy; lipid; membrane protein; molecular dynamics; simulation.
© 2019 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures

Similar articles
-
The importance of membrane defects-lessons from simulations.Acc Chem Res. 2014 Aug 19;47(8):2244-51. doi: 10.1021/ar4002729. Epub 2014 Jun 3. Acc Chem Res. 2014. PMID: 24892900
-
Association of Peripheral Membrane Proteins with Membranes: Free Energy of Binding of GRP1 PH Domain with Phosphatidylinositol Phosphate-Containing Model Bilayers.J Phys Chem Lett. 2016 Apr 7;7(7):1219-24. doi: 10.1021/acs.jpclett.6b00153. Epub 2016 Mar 17. J Phys Chem Lett. 2016. PMID: 26977543 Free PMC article.
-
Interactions of Pleckstrin Homology Domains with Membranes: Adding Back the Bilayer via High-Throughput Molecular Dynamics.Structure. 2016 Aug 2;24(8):1421-1431. doi: 10.1016/j.str.2016.06.002. Epub 2016 Jul 14. Structure. 2016. PMID: 27427480 Free PMC article.
-
Molecular dynamics simulations of GPCR-cholesterol interaction: An emerging paradigm.Biochim Biophys Acta. 2015 Sep;1848(9):1775-82. doi: 10.1016/j.bbamem.2015.03.018. Epub 2015 Mar 25. Biochim Biophys Acta. 2015. PMID: 25817549 Review.
-
The influence of cholesterol on membrane protein structure, function, and dynamics studied by molecular dynamics simulations.Biochim Biophys Acta. 2015 Sep;1848(9):1783-95. doi: 10.1016/j.bbamem.2015.03.029. Epub 2015 Apr 1. Biochim Biophys Acta. 2015. PMID: 25839353 Review.
Cited by
-
The transport pathway in the ABCG2 protein and its regulation revealed by molecular dynamics simulations.Cell Mol Life Sci. 2021 Mar;78(5):2329-2339. doi: 10.1007/s00018-020-03651-3. Epub 2020 Sep 26. Cell Mol Life Sci. 2021. PMID: 32979053 Free PMC article.
-
Protein-mediated interactions in the dynamic regulation of acute inflammation.Biocell. 2023;47(6):1191-1198. doi: 10.32604/biocell.2023.027838. Epub 2023 May 19. Biocell. 2023. PMID: 37261220 Free PMC article.
-
Estimating the Cholesterol Affinity of Integral Membrane Proteins from Experimental Data.Biochemistry. 2024 Jan 2;63(1):19-26. doi: 10.1021/acs.biochem.3c00567. Epub 2023 Dec 15. Biochemistry. 2024. PMID: 38099740 Free PMC article.
-
From Bench to Biomolecular Simulation: Phospholipid Modulation of Potassium Channels.J Mol Biol. 2021 Aug 20;433(17):167105. doi: 10.1016/j.jmb.2021.167105. Epub 2021 Jun 15. J Mol Biol. 2021. PMID: 34139216 Free PMC article. Review.
-
Structural Basis of PE_PGRS Polymorphism, a Tool for Functional Modulation.Biomolecules. 2023 May 10;13(5):812. doi: 10.3390/biom13050812. Biomolecules. 2023. PMID: 37238682 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- BB/R00126X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- BB/R002517/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/P01948X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 208361/Z/17/Z/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

