CDK1 and CCNB1 as potential diagnostic markers of rhabdomyosarcoma: validation following bioinformatics analysis
- PMID: 31870357
- PMCID: PMC6929508
- DOI: 10.1186/s12920-019-0645-x
CDK1 and CCNB1 as potential diagnostic markers of rhabdomyosarcoma: validation following bioinformatics analysis
Abstract
Background: Rhabdomyosarcoma (RMS), a common soft-tissue malignancy in pediatrics, presents high invasiveness and mortality. However, besides known changes in the PAX3/7-FOXO1 fusion gene in alveolar RMS, the molecular mechanisms of the disease remain incompletely understood. The purpose of the study is to recognize potential biomarkers related with RMS and analyse their molecular mechanism, diagnosis and prognostic significance.
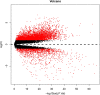
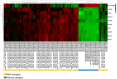
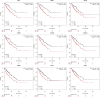
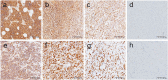
Methods: The Gene Expression Omnibus was used to search the RMS and normal striated muscle data sets. Differentially expressed genes (DEGs) were filtered using R software. The DAVID has become accustomed to performing functional annotations and pathway analysis on DEGs. The protein interaction was constructed and further processed by the STRING tool and Cytoscape software. Kaplan-Meier was used to estimate the effect of hub genes on the ending of sarcoma sufferers, and the expression of these genes in RMS was proved by real-time polymerase chain reaction (RT-PCR). Finally, the expression of CDK1 and CCNB1 in RMS was validated by immunohistochemistry (IHC).
Results: A total of 1932 DEGs were obtained, amongst which 1505 were up-regulated and 427were down-regulated. Up-regulated genes were largely enriched in the cell cycle, ECM-receptor interaction, PI3K/Akt and p53 pathways, whilst down-regulated genes were primarily enriched in the muscle contraction process. CDK1, CCNB1, CDC20, CCNB2, AURKB, MAD2L1, HIST2H2BE, CENPE, KIF2C and PCNA were identified as hub genes by Cytoscape analyses. Survival analysis showed that, except for HIST2H2BE, the other hub genes were highly expressed and related to poor prognosis in sarcoma. RT-PCR validation showed that CDK1, CCNB1, CDC20, CENPE and HIST2H2BE were significantly differential expression in RMS compared to the normal control. IHC revealed that the expression of CDK1 (28/32, 87.5%) and CCNB1 (26/32, 81.25%) were notably higher in RMS than normal controls (1/9, 11.1%; 0/9, 0%). Moreover, the CCNB1 was associated with the age and location of the patient's onset.
Conclusions: These results show that these hub genes, especially CDK1 and CCNB1, may be potential diagnostic biomarkers for RMS and provide a new perspective for the pathogenesis of RMS.
Keywords: Bioinformatics analysis; CCNB1; CDK1; Diagnosis; Hub genes; Rhabdomyosarcoma.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
Upregulated cyclins may be novel genes for triple-negative breast cancer based on bioinformatic analysis.Breast Cancer. 2020 Sep;27(5):903-911. doi: 10.1007/s12282-020-01086-z. Epub 2020 Apr 27. Breast Cancer. 2020. PMID: 32338339
-
Identification of Multiple Hub Genes and Pathways in Hepatocellular Carcinoma: A Bioinformatics Analysis.Biomed Res Int. 2021 Jul 12;2021:8849415. doi: 10.1155/2021/8849415. eCollection 2021. Biomed Res Int. 2021. PMID: 34337056 Free PMC article.
-
Screening Hub Genes as Prognostic Biomarkers of Hepatocellular Carcinoma by Bioinformatics Analysis.Cell Transplant. 2019 Dec;28(1_suppl):76S-86S. doi: 10.1177/0963689719893950. Epub 2019 Dec 11. Cell Transplant. 2019. PMID: 31822116 Free PMC article.
-
Molecular diagnostics in the management of rhabdomyosarcoma.Expert Rev Mol Diagn. 2017 Feb;17(2):189-194. doi: 10.1080/14737159.2017.1275965. Epub 2017 Jan 6. Expert Rev Mol Diagn. 2017. PMID: 28058850 Free PMC article. Review.
-
Transcriptomic Signatures in Colorectal Cancer Progression.Curr Mol Med. 2023;23(3):239-249. doi: 10.2174/1566524022666220427102048. Curr Mol Med. 2023. PMID: 35490318 Review.
Cited by
-
DCX and CRABP2 are candidate genes for differential diagnosis between pre-chemotherapy embryonic and alveolar rhabdomyosarcoma in pediatric patients.Pediatr Investig. 2021 Jun 18;5(2):106-111. doi: 10.1002/ped4.12278. eCollection 2021 Jun. Pediatr Investig. 2021. PMID: 34179706 Free PMC article.
-
Evolutionary Perspective and Expression Analysis of Intronless Genes Highlight the Conservation of Their Regulatory Role.Front Genet. 2021 Jul 9;12:654256. doi: 10.3389/fgene.2021.654256. eCollection 2021. Front Genet. 2021. PMID: 34306008 Free PMC article.
-
CCNB1 is a novel prognostic biomarker and promotes proliferation, migration and invasion in Wilms tumor.BMC Med Genomics. 2023 Aug 17;16(1):189. doi: 10.1186/s12920-023-01627-3. BMC Med Genomics. 2023. PMID: 37592341 Free PMC article.
-
MYBPC2 and MYL1 as Significant Gene Markers for Rhabdomyosarcoma.Technol Cancer Res Treat. 2021 Jan-Dec;20:1533033820979669. doi: 10.1177/1533033820979669. Technol Cancer Res Treat. 2021. PMID: 33499774 Free PMC article.
-
Cyclin-dependent kinases: Masters of the eukaryotic universe.Wiley Interdiscip Rev RNA. 2023 Sep 17;15(1):e1816. doi: 10.1002/wrna.1816. Online ahead of print. Wiley Interdiscip Rev RNA. 2023. PMID: 37718413 Free PMC article. Review.
References
-
- Davicioni E, Anderson MJ, Finckenstein FG, Lynch JC, Qualman SJ, Shimada H, Schofield DE, Buckley JD, Meyer WH, Sorensen PH, et al. Molecular classification of rhabdomyosarcoma--genotypic and phenotypic determinants of diagnosis: a report from the Children's oncology group. Am J Pathol. 2009;174(2):550–564. doi: 10.2353/ajpath.2009.080631. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

