Identification of HCMV-derived T cell epitopes in seropositive individuals through viral deletion models
- PMID: 31869419
- PMCID: PMC7062530
- DOI: 10.1084/jem.20191164
Identification of HCMV-derived T cell epitopes in seropositive individuals through viral deletion models
Abstract
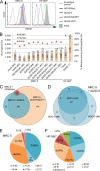
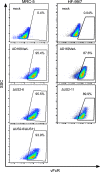
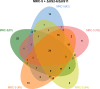
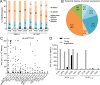
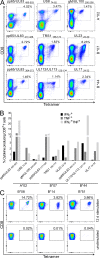
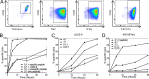
In healthy individuals, immune control of persistent human cytomegalovirus (HCMV) infection is effectively mediated by virus-specific CD4+ and CD8+ T cells. However, identifying the repertoire of T cell specificities for HCMV is hampered by the immense protein coding capacity of this betaherpesvirus. Here, we present a novel approach that employs HCMV deletion mutant viruses lacking HLA class I immunoevasins and allows direct identification of naturally presented HCMV-derived HLA ligands by mass spectrometry. We identified 368 unique HCMV-derived HLA class I ligands representing an unexpectedly broad panel of 123 HCMV antigens. Functional characterization revealed memory T cell responses in seropositive individuals for a substantial proportion (28%) of these novel peptides. Multiple HCMV-directed specificities in the memory T cell pool of single individuals indicate that physiologic anti-HCMV T cell responses are directed against a broad range of antigens. Thus, the unbiased identification of naturally presented viral epitopes enabled a comprehensive and systematic assessment of the physiological repertoire of anti-HCMV T cell specificities in seropositive individuals.
© 2019 Lübke et al.
Conflict of interest statement
Disclosures: M. Lübke reported a patent to HCMV epitopes (pending). S. Spalt reported a patent to HCMV epitopes (pending) and is currently an employee of Immatics Biotechnologies GmbH. D.J. Kowalewski reported a patent to HCMV epitopes (pending) and is currently an employee of Immatics Biotechnologies. C. Zimmermann reported a patent to HCMV epitopes (pending). L. Bauersfeld reported a patent to HCMV epitopes (pending). A. Nelde reported a patent to HCMV epitopes (pending). V.T.K. Le-Trilling reported a patent to HCMV epitopes (pending). H. Hengel reported a patent to HCMV epitopes (pending). H.-G. Rammensee reported "other" from Immatics GmbH and "other" from Curevac AG during the conduct of the study; in addition, H.-G. Rammensee had a patent to HCMV epitopes (pending). A. Halenius reported a patent to HCMV epitopes (pending). No other disclosures were reported.
Figures



Similar articles
-
Human Macrophages Escape Inhibition of Major Histocompatibility Complex-Dependent Antigen Presentation by Cytomegalovirus and Drive Proliferation and Activation of Memory CD4+ and CD8+ T Cells.Front Immunol. 2018 May 25;9:1129. doi: 10.3389/fimmu.2018.01129. eCollection 2018. Front Immunol. 2018. PMID: 29887865 Free PMC article.
-
Ex vivo profiling of CD8+-T-cell responses to human cytomegalovirus reveals broad and multispecific reactivities in healthy virus carriers.J Virol. 2003 May;77(9):5226-40. doi: 10.1128/jvi.77.9.5226-5240.2003. J Virol. 2003. PMID: 12692225 Free PMC article.
-
Presentation of an immunodominant immediate-early CD8+ T cell epitope resists human cytomegalovirus immunoevasion.PLoS Pathog. 2013;9(5):e1003383. doi: 10.1371/journal.ppat.1003383. Epub 2013 May 23. PLoS Pathog. 2013. PMID: 23717207 Free PMC article. Clinical Trial.
-
Generation, maintenance and tissue distribution of T cell responses to human cytomegalovirus in lytic and latent infection.Med Microbiol Immunol. 2019 Aug;208(3-4):375-389. doi: 10.1007/s00430-019-00598-6. Epub 2019 Mar 20. Med Microbiol Immunol. 2019. PMID: 30895366 Free PMC article. Review.
-
Molecular characterization of HCMV-specific immune responses: Parallels between CD8(+) T cells, CD4(+) T cells, and NK cells.Eur J Immunol. 2015 Sep;45(9):2433-45. doi: 10.1002/eji.201545495. Epub 2015 Aug 24. Eur J Immunol. 2015. PMID: 26228786 Review.
Cited by
-
T Cell Epitope Discovery in the Context of Distinct and Unique Indigenous HLA Profiles.Front Immunol. 2022 May 6;13:812393. doi: 10.3389/fimmu.2022.812393. eCollection 2022. Front Immunol. 2022. PMID: 35603215 Free PMC article. Review.
-
SARS-CoV-2-derived peptides define heterologous and COVID-19-induced T cell recognition.Nat Immunol. 2021 Jan;22(1):74-85. doi: 10.1038/s41590-020-00808-x. Epub 2020 Sep 30. Nat Immunol. 2021. PMID: 32999467
-
Orf Virus-Based Vaccine Vector D1701-V Induces Strong CD8+ T Cell Response against the Transgene but Not against ORFV-Derived Epitopes.Vaccines (Basel). 2020 Jun 10;8(2):295. doi: 10.3390/vaccines8020295. Vaccines (Basel). 2020. PMID: 32531997 Free PMC article.
-
Solving an MHC allele-specific bias in the reported immunopeptidome.JCI Insight. 2020 Oct 2;5(19):e141264. doi: 10.1172/jci.insight.141264. JCI Insight. 2020. PMID: 32897882 Free PMC article.
-
Prevalent and immunodominant CD8 T cell epitopes are conserved in SARS-CoV-2 variants.Cell Rep. 2023 Jan 31;42(1):111995. doi: 10.1016/j.celrep.2023.111995. Epub 2023 Jan 9. Cell Rep. 2023. PMID: 36656713 Free PMC article.
References
-
- Britten C.M., Gouttefangeas C., Welters M.J., Pawelec G., Koch S., Ottensmeier C., Mander A., Walter S., Paschen A., Müller-Berghaus J., et al. . 2008. The CIMT-monitoring panel: a two-step approach to harmonize the enumeration of antigen-specific CD8+ T lymphocytes by structural and functional assays. Cancer Immunol. Immunother. 57:289–302. 10.1007/s00262-007-0378-0 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

