The HIV-Tat protein interacts with Sp3 transcription factor and inhibits its binding to a distal site of the sod2 promoter in human pulmonary artery endothelial cells
- PMID: 31863909
- PMCID: PMC7039131
- DOI: 10.1016/j.freeradbiomed.2019.12.015
The HIV-Tat protein interacts with Sp3 transcription factor and inhibits its binding to a distal site of the sod2 promoter in human pulmonary artery endothelial cells
Abstract
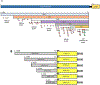
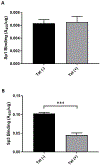
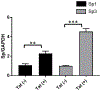
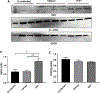
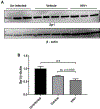
Redox imbalance results in damage to cellular macromolecules and interferes with signaling pathways, leading to an inflammatory cellular and tissue environment. As such, the cellular oxidative environment is tightly regulated by several redox-modulating pathways. Many viruses have evolved intricate mechanisms to manipulate these pathways for their benefit, including HIV-1, which requires a pro-oxidant cellular environment for optimal replication. One such virulence factor responsible for modulating the redox environment is the HIV Transactivator of transcription (Tat). Tat is of particular interest as it is actively secreted by infected cells and internalized by uninfected bystander cells where it can elicit pro-oxidant effects resulting in inflammation and damage. Previously, we demonstrated that Tat regulates basal expression of Superoxide Dismutase 2 (sod2) by altering the binding of the Sp-transcription factors at regions relatively near (approx. -210 nucleotides) upstream of the transcriptional start site. Now, using in silico analysis and a series of sod2 promoter reporter constructs, we have identified putative clusters of Sp-binding sites located further upstream of the proximal sod2 promoter, between nucleotides -3400 to -210, and tested their effect on basal transcription and for their sensitivity to HIV-1 Tat. In this report, we demonstrate that under basal conditions, maximal transcription requires a cluster of Sp-binding sites in the -584 nucleotide region, which is extremely sensitive to Tat. Using chromatin immunoprecipitation (ChIP) we demonstrate that Tat results in altered occupancy of Sp1 and Sp3 at this distal Tat-sensitive regulatory element and strongly stimulated endogenous expression of SOD2 in human pulmonary artery endothelial cells (HPAEC). We also report altered expression of Sp1 and Sp3 in Tat-expressing HPAEC as well as in the lungs of HIV-1 infected humanized mice. Lastly, Tat co-immunoprecipitated with endogenous Sp3 but not Sp1 and did not alter the acetylation state of Sp3. Thus, here, we have defined a novel and important cis-acting factor in HIV-1 Tat-mediated regulation of SOD2, demonstrated that modulation of Sp1 and Sp3 activity by Tat promotes SOD2 expression in primary human pulmonary artery endothelial cells and determined that pulmonary levels of Sp3 as well as SOD2 are increased in the lungs of a mouse model of HIV infection.
Published by Elsevier Inc.
Figures

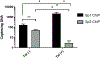


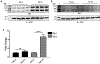

Similar articles
-
HIV-1 Tat regulates the SOD2 basal promoter by altering Sp1/Sp3 binding activity.Free Radic Biol Med. 2004 Sep 15;37(6):869-80. doi: 10.1016/j.freeradbiomed.2004.06.016. Free Radic Biol Med. 2004. PMID: 15706661
-
Different roles of Sp family members in HIV-1 Tat-mediated manganese superoxide dismutase suppression in hepatocellular carcinoma cells.DNA Cell Biol. 2005 May;24(5):299-310. doi: 10.1089/dna.2005.24.299. DNA Cell Biol. 2005. PMID: 15869407
-
Transcription factors sp1 and sp3 regulate expression of human extracellular superoxide dismutase in lung fibroblasts.Am J Respir Cell Mol Biol. 2008 Aug;39(2):243-51. doi: 10.1165/rcmb.2007-0378OC. Epub 2008 Feb 28. Am J Respir Cell Mol Biol. 2008. PMID: 18314536 Free PMC article.
-
Sp1 and Sp3 transcription factors regulate the basal expression of human microsomal epoxide hydrolase (EPHX1) through interaction with the E1b far upstream promoter.Gene. 2014 Feb 15;536(1):135-44. doi: 10.1016/j.gene.2013.11.053. Epub 2013 Dec 4. Gene. 2014. PMID: 24315822 Free PMC article.
-
Defining the molecular mechanisms of HIV-1 Tat secretion: PtdIns(4,5)P2 at the epicenter.Traffic. 2018 Apr 30:10.1111/tra.12578. doi: 10.1111/tra.12578. Online ahead of print. Traffic. 2018. PMID: 29708629 Free PMC article. Review.
Cited by
-
The DNA Damage Response and HIV-Associated Pulmonary Arterial Hypertension.Int J Mol Sci. 2020 May 7;21(9):3305. doi: 10.3390/ijms21093305. Int J Mol Sci. 2020. PMID: 32392789 Free PMC article. Review.
-
The Intersection of HIV and Pulmonary Vascular Health: From HIV Evolution to Vascular Cell Types to Disease Mechanisms.J Vasc Dis. 2024 Jun;3(2):174-200. doi: 10.3390/jvd3020015. Epub 2024 May 6. J Vasc Dis. 2024. PMID: 39464800 Free PMC article.
References
-
- Marciniak RA, Calnan BJ, Frankel AD, Sharp PA, HIV-1 Tat Protein Tram-Activates Transcription In Vitro, Cell. 63 (1990) 791–602. http://ac.els-cdn.com.hsl-ezproxy.ucdenver.edu/0092867490901455/1-s2.0-0... (accessed April 25, 2017). - PubMed
-
- Marciniak RA, Sharp PA, HIV-1 Tat protein promotes formation of more-processive elongation complexes, EMBO J. 1013 (1991) 4189–4196. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC453171/pdf/emboj00111-0201.pdf (accessed April 25, 2017). - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

