When RAD52 Allows Mitosis to Accept Unscheduled DNA Synthesis
- PMID: 31861741
- PMCID: PMC7017103
- DOI: 10.3390/cancers12010026
When RAD52 Allows Mitosis to Accept Unscheduled DNA Synthesis
Abstract
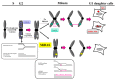
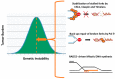
Faithful duplication of the human genome during the S phase of cell cycle and accurate segregation of sister chromatids in mitosis are essential for the maintenance of chromosome stability from one generation of cells to the next. Cells that are copying their DNA in preparation for division can suffer from 'replication stress' (RS) due to various external or endogenous impediments that slow or stall replication forks. RS is a major cause of pathologies including cancer, premature ageing and other disorders associated with genomic instability. It particularly affects genomic loci where progression of replication forks is intrinsically slow or problematic, such as common fragile site (CFS), telomeres, and repetitive sequences. Although the eukaryotic cell cycle is conventionally thought of as several separate steps, each of which must be completed before the next one is initiated, it is now accepted that incompletely replicated chromosomal domains generated in S phase upon RS at these genomic loci can result in late DNA synthesis in G2/M. In 2013, during investigations into the mechanism by which the specialized DNA polymerase eta (Pol η) contributes to the replication and stability of CFS, we unveiled that indeed some DNA synthesis was still occurring in early mitosis at these loci. This surprising observation of mitotic DNA synthesis that differs fundamentally from canonical semi-conservative DNA replication in S-phase has been then confirmed, called "MiDAS"and believed to counteract potentially lethal chromosome mis-segregation and non-disjunction. While other contributions in this Special Issue of Cancers focus on the role of RAS52RAD52 during MiDAS, this review emphases on the discovery of MiDAS and its molecular effectors.
Keywords: DNA replication; RAD52; chromosome instability; genome instability; mitotic DNA synthesis; replication stress.
Conflict of interest statement
The authors have no affiliations or financial involvement with any entity with a financial interest or conflict with the material in this review.
Figures

Similar articles
-
Mitotic DNA Synthesis in Untransformed Human Cells Preserves Common Fragile Site Stability via a FANCD2-Driven Mechanism That Requires HELQ.J Mol Biol. 2023 Nov 15;435(22):168294. doi: 10.1016/j.jmb.2023.168294. Epub 2023 Sep 28. J Mol Biol. 2023. PMID: 37777152 Free PMC article.
-
RAD52 Facilitates Mitotic DNA Synthesis Following Replication Stress.Mol Cell. 2016 Dec 15;64(6):1117-1126. doi: 10.1016/j.molcel.2016.10.037. Mol Cell. 2016. PMID: 27984745
-
Mind the replication gap.R Soc Open Sci. 2021 Jun 9;8(6):201932. doi: 10.1098/rsos.201932. R Soc Open Sci. 2021. PMID: 34113447 Free PMC article. Review.
-
Inducing and Detecting Mitotic DNA Synthesis at Difficult-to-Replicate Loci.Methods Enzymol. 2018;601:45-58. doi: 10.1016/bs.mie.2017.11.025. Epub 2018 Feb 3. Methods Enzymol. 2018. PMID: 29523241 Review.
-
Human cancer cells utilize mitotic DNA synthesis to resist replication stress at telomeres regardless of their telomere maintenance mechanism.Oncotarget. 2018 Mar 23;9(22):15836-15846. doi: 10.18632/oncotarget.24745. eCollection 2018 Mar 23. Oncotarget. 2018. PMID: 29662610 Free PMC article.
Cited by
-
Structural Chromosome Instability: Types, Origins, Consequences, and Therapeutic Opportunities.Cancers (Basel). 2021 Jun 19;13(12):3056. doi: 10.3390/cancers13123056. Cancers (Basel). 2021. PMID: 34205328 Free PMC article. Review.
-
53BP1: Keeping It under Control, Even at a Distance from DNA Damage.Genes (Basel). 2022 Dec 16;13(12):2390. doi: 10.3390/genes13122390. Genes (Basel). 2022. PMID: 36553657 Free PMC article. Review.
-
Regulator of telomere elongation helicase 1 gene and its association with malignancy.Cancer Rep (Hoboken). 2023 Jan;6(1):e1735. doi: 10.1002/cnr2.1735. Epub 2022 Oct 17. Cancer Rep (Hoboken). 2023. PMID: 36253342 Free PMC article. Review.
-
Current Understanding of RAD52 Functions: Fundamental and Therapeutic Insights.Cancers (Basel). 2020 Mar 17;12(3):705. doi: 10.3390/cancers12030705. Cancers (Basel). 2020. PMID: 32192055 Free PMC article.
-
Processing DNA lesions during mitosis to prevent genomic instability.Biochem Soc Trans. 2022 Aug 31;50(4):1105-1118. doi: 10.1042/BST20220049. Biochem Soc Trans. 2022. PMID: 36040211 Free PMC article. Review.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

