Epigenetic Control of Autophagy in Cancer Cells: A Key Process for Cancer-Related Phenotypes
- PMID: 31861179
- PMCID: PMC6952790
- DOI: 10.3390/cells8121656
Epigenetic Control of Autophagy in Cancer Cells: A Key Process for Cancer-Related Phenotypes
Abstract
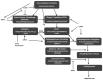
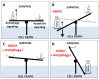
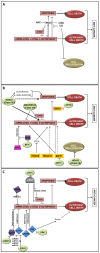
Although autophagy is a well-known and extensively described cell pathway, numerous studies have been recently interested in studying the importance of its regulation at different molecular levels, including the translational and post-translational levels. Therefore, this review focuses on the links between autophagy and epigenetics in cancer and summarizes the. following: (i) how ATG genes are regulated by epigenetics, including DNA methylation and post-translational histone modifications; (ii) how epidrugs are able to modulate autophagy in cancer and to alter cancer-related phenotypes (proliferation, migration, invasion, tumorigenesis, etc.) and; (iii) how epigenetic enzymes can also regulate autophagy at the protein level. One noteable observation was that researchers most often reported conclusions about the regulation of the autophagy flux, following the use of epidrugs, based only on the analysis of LC3B-II form in treated cells. However, it is now widely accepted that an increase in LC3B-II form could be the consequence of an induction of the autophagy flux, as well as a block in the autophagosome-lysosome fusion. Therefore, in our review, all the published results describing a link between epidrugs and autophagy were systematically reanalyzed to determine whether autophagy flux was indeed increased, or inhibited, following the use of these potentially new interesting treatments targeting the autophagy process. Altogether, these recent data strongly support the idea that the determination of autophagy status could be crucial for future anticancer therapies. Indeed, the use of a combination of epidrugs and autophagy inhibitors could be beneficial for some cancer patients, whereas, in other cases, an increase of autophagy, which is frequently observed following the use of epidrugs, could lead to increased autophagy cell death.
Keywords: DNA methylation; autophagy; cancer; epigenetics; histone deacetylase (HDAC); histone methylation.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Epigenetic Targeting of Autophagy via HDAC Inhibition in Tumor Cells: Role of p53.Int J Mol Sci. 2018 Dec 8;19(12):3952. doi: 10.3390/ijms19123952. Int J Mol Sci. 2018. PMID: 30544838 Free PMC article. Review.
-
Dysregulation of histone deacetylases in carcinogenesis and tumor progression: a possible link to apoptosis and autophagy.Cell Mol Life Sci. 2019 Sep;76(17):3263-3282. doi: 10.1007/s00018-019-03098-1. Epub 2019 Apr 13. Cell Mol Life Sci. 2019. PMID: 30982077 Free PMC article. Review.
-
Epigenetic therapy of cancer with histone deacetylase inhibitors.J Cancer Res Ther. 2014 Jul-Sep;10(3):469-78. doi: 10.4103/0973-1482.137937. J Cancer Res Ther. 2014. PMID: 25313724 Review.
-
LC3B-II deacetylation by histone deacetylase 6 is involved in serum-starvation-induced autophagic degradation.Biochem Biophys Res Commun. 2013 Nov 29;441(4):970-5. doi: 10.1016/j.bbrc.2013.11.007. Epub 2013 Nov 9. Biochem Biophys Res Commun. 2013. PMID: 24220335
-
Research Advances in the Use of Histone Deacetylase Inhibitors for Epigenetic Targeting of Cancer.Curr Top Med Chem. 2019;19(12):995-1004. doi: 10.2174/1568026619666190125145110. Curr Top Med Chem. 2019. PMID: 30686256 Review.
Cited by
-
Melatonin inhibits tongue squamous cell carcinoma: Interplay of ER stress-induced apoptosis and autophagy with cell migration.Heliyon. 2024 Apr 9;10(8):e29291. doi: 10.1016/j.heliyon.2024.e29291. eCollection 2024 Apr 30. Heliyon. 2024. PMID: 38644851 Free PMC article.
-
The Role of LncRNAs in Uveal Melanoma.Cancers (Basel). 2021 Aug 11;13(16):4041. doi: 10.3390/cancers13164041. Cancers (Basel). 2021. PMID: 34439196 Free PMC article. Review.
-
Epigenetic Priming with Decitabine Augments the Therapeutic Effect of Cisplatin on Triple-Negative Breast Cancer Cells through Induction of Proapoptotic Factor NOXA.Cancers (Basel). 2022 Jan 4;14(1):248. doi: 10.3390/cancers14010248. Cancers (Basel). 2022. PMID: 35008411 Free PMC article.
-
All-Trans-Retinoic Acid Combined With Valproic Acid Can Promote Differentiation in Myeloid Leukemia Cells by an Autophagy Dependent Mechanism.Front Oncol. 2022 Feb 24;12:848517. doi: 10.3389/fonc.2022.848517. eCollection 2022. Front Oncol. 2022. PMID: 35280824 Free PMC article.
-
Autophagy-Dependent Sensitization of Triple-Negative Breast Cancer Models to Topoisomerase II Poisons by Inhibition of the Nucleosome Remodeling Factor.Mol Cancer Res. 2021 Aug;19(8):1338-1349. doi: 10.1158/1541-7786.MCR-20-0743. Epub 2021 Apr 2. Mol Cancer Res. 2021. PMID: 33811160 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

