Lessons from eRNAs: understanding transcriptional regulation through the lens of nascent RNAs
- PMID: 31856658
- PMCID: PMC7053884
- DOI: 10.1080/21541264.2019.1704128
Lessons from eRNAs: understanding transcriptional regulation through the lens of nascent RNAs
Abstract
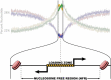
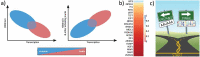
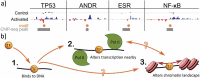
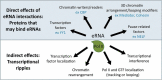
Nascent transcription assays, such as global run-on sequencing (GRO-seq) and precision run-on sequencing (PRO-seq), have uncovered a myriad of unstable RNAs being actively produced from numerous sites genome-wide. These transcripts provide a more complete and immediate picture of the impact of regulatory events. Transcription factors recruit RNA polymerase II, effectively initiating the process of transcription; repressors inhibit polymerase recruitment. Efficiency of recruitment is dictated by sequence elements in and around the RNA polymerase loading zone. A combination of sequence elements and RNA binding proteins subsequently influence the ultimate stability of the resulting transcript. Some of these transcripts are capable of providing feedback on the process, influencing subsequent transcription. By monitoring RNA polymerase activity, nascent assays provide insights into every step of the regulated process of transcription.
Keywords: RNA polymerase II; chromatin structure; eRNAs; enhancers; gene expression; nascent RNA; non-coding RNA function; transcription factors.
Figures
Similar articles
-
Computational Approaches for Mining GRO-Seq Data to Identify and Characterize Active Enhancers.Methods Mol Biol. 2017;1468:121-38. doi: 10.1007/978-1-4939-4035-6_10. Methods Mol Biol. 2017. PMID: 27662874 Free PMC article.
-
Enhancer RNAs step forward: new insights into enhancer function.Development. 2022 Aug 15;149(16):dev200398. doi: 10.1242/dev.200398. Epub 2022 Aug 30. Development. 2022. PMID: 36039999 Free PMC article.
-
Enhancer RNAs: a missing regulatory layer in gene transcription.Sci China Life Sci. 2019 Jul;62(7):905-912. doi: 10.1007/s11427-017-9370-9. Epub 2018 Dec 26. Sci China Life Sci. 2019. PMID: 30593613 Review.
-
RNA Targets Ribogenesis Factor WDR43 to Chromatin for Transcription and Pluripotency Control.Mol Cell. 2019 Jul 11;75(1):102-116.e9. doi: 10.1016/j.molcel.2019.05.007. Epub 2019 May 22. Mol Cell. 2019. PMID: 31128943
-
Enhancer RNA: biogenesis, function, and regulation.Essays Biochem. 2020 Dec 7;64(6):883-894. doi: 10.1042/EBC20200014. Essays Biochem. 2020. PMID: 33034351 Review.
Cited by
-
A retrotransposon storm marks clinical phenoconversion to late-onset Alzheimer's disease.Geroscience. 2022 Jun;44(3):1525-1550. doi: 10.1007/s11357-022-00580-w. Epub 2022 May 19. Geroscience. 2022. PMID: 35585302 Free PMC article.
-
m6A readers, writers, erasers, and the m6A epitranscriptome in breast cancer.J Mol Endocrinol. 2022 Dec 21;70(2):e220110. doi: 10.1530/JME-22-0110. Print 2023 Feb 1. J Mol Endocrinol. 2022. PMID: 36367225 Free PMC article. Review.
-
Stress-induced transcriptional memory accelerates promoter-proximal pause release and decelerates termination over mitotic divisions.Mol Cell. 2021 Apr 15;81(8):1715-1731.e6. doi: 10.1016/j.molcel.2021.03.007. Epub 2021 Mar 29. Mol Cell. 2021. PMID: 33784494 Free PMC article.
-
Protocol variations in run-on transcription dataset preparation produce detectable signatures in sequencing libraries.BMC Genomics. 2022 Mar 7;23(1):187. doi: 10.1186/s12864-022-08352-8. BMC Genomics. 2022. PMID: 35255806 Free PMC article.
-
The Mediator complex as a master regulator of transcription by RNA polymerase II.Nat Rev Mol Cell Biol. 2022 Nov;23(11):732-749. doi: 10.1038/s41580-022-00498-3. Epub 2022 Jun 20. Nat Rev Mol Cell Biol. 2022. PMID: 35725906 Free PMC article. Review.
References
-
- Mifsud B, Tavares-Cadete F, Young AN, et al. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat Genet. 2015;47(6):598–606. - PubMed
-
- Shlyueva D, Stampfel G, Stark A.. Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet. 2014;15(4):272–286. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
