Novel Reassortant Highly Pathogenic Avian Influenza A(H5N2) Virus in Broiler Chickens, Egypt
- PMID: 31855539
- PMCID: PMC6924912
- DOI: 10.3201/eid2601.190570
Novel Reassortant Highly Pathogenic Avian Influenza A(H5N2) Virus in Broiler Chickens, Egypt
Abstract
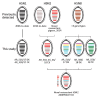
We detected a novel reassortant highly pathogenic avian influenza A(H5N2) virus in 3 poultry farms in Egypt. The virus carried genome segments of a pigeon H9N2 influenza virus detected in 2014, a nucleoprotein segment of contemporary chicken H9N2 viruses from Egypt, and hemagglutinin derived from the 2.3.4.4b H5N8 virus clade.
Keywords: Egypt; H5N2; avian influenza virus; broiler chickens; highly pathogenic avian influenza; influenza; poultry; reassortant; respiratory infections; viruses; zoonoses.
Figures

Similar articles
-
Genotyping and reassortment analysis of highly pathogenic avian influenza viruses H5N8 and H5N2 from Egypt reveals successive annual replacement of genotypes.Infect Genet Evol. 2020 Oct;84:104375. doi: 10.1016/j.meegid.2020.104375. Epub 2020 May 23. Infect Genet Evol. 2020. PMID: 32454245
-
Emergence of a novel reassortant highly pathogenic avian influenza clade 2.3.4.4b A(H5N2) Virus, 2024.Emerg Microbes Infect. 2025 Dec;14(1):2455601. doi: 10.1080/22221751.2025.2455601. Epub 2025 Jan 31. Emerg Microbes Infect. 2025. PMID: 39868968 Free PMC article.
-
Genetic and biological characterization of two reassortant H5N2 avian influenza A viruses isolated from waterfowl in China in 2016.Vet Microbiol. 2018 Oct;224:8-16. doi: 10.1016/j.vetmic.2018.08.016. Epub 2018 Aug 15. Vet Microbiol. 2018. PMID: 30269795
-
Pandemic potential of highly pathogenic avian influenza clade 2.3.4.4 A(H5) viruses.Rev Med Virol. 2020 May;30(3):e2099. doi: 10.1002/rmv.2099. Epub 2020 Mar 5. Rev Med Virol. 2020. PMID: 32135031 Free PMC article. Review.
-
A Review of the Stability of Avian Influenza Virus in Materials from Poultry Farms.Avian Dis. 2023 Sep;67(3):229-236. doi: 10.1637/aviandiseases-D-23-00027. Avian Dis. 2023. PMID: 39126409 Review.
Cited by
-
Genetic incompatibilities and reduced transmission in chickens may limit the evolution of reassortants between H9N2 and panzootic H5N8 clade 2.3.4.4 avian influenza virus showing high virulence for mammals.Virus Evol. 2020 Oct 15;6(2):veaa077. doi: 10.1093/ve/veaa077. eCollection 2020 Jul. Virus Evol. 2020. PMID: 33343923 Free PMC article.
-
Molecular characterization of the whole genome of H9N2 avian influenza virus isolated from Egyptian poultry farms.Arch Virol. 2024 Apr 16;169(5):99. doi: 10.1007/s00705-024-06018-2. Arch Virol. 2024. PMID: 38625394 Free PMC article.
-
Genetic Characterization and Pathogenesis of Three Novel Reassortant H5N2 Viruses in South Korea, 2018.Viruses. 2021 Oct 30;13(11):2192. doi: 10.3390/v13112192. Viruses. 2021. PMID: 34834997 Free PMC article.
-
Genetic Variations among Different Variants of G1-like Avian Influenza H9N2 Viruses and Their Pathogenicity in Chickens.Viruses. 2022 May 11;14(5):1030. doi: 10.3390/v14051030. Viruses. 2022. PMID: 35632771 Free PMC article.
-
Genetic and Antigenic Characteristics of Highly Pathogenic Avian Influenza A(H5N8) Viruses Circulating in Domestic Poultry in Egypt, 2017-2021.Microorganisms. 2022 Mar 9;10(3):595. doi: 10.3390/microorganisms10030595. Microorganisms. 2022. PMID: 35336170 Free PMC article.
References
-
- World Health Organization. Cumulative number of confirmed human cases for avian influenza A(H5N1) reported to WHO, 2003–2019. 2019. [cited 2019 Mar 4]. https://www.who.int/influenza/human_animal_interface/2019_02_12_tableH5N...
-
- Naguib MM, Ulrich R, Kasbohm E, Eng CLP, Hoffmann D, Grund C, et al. Natural reassortants of potentially zoonotic avian influenza viruses H5N1 and H9N2 from Egypt display distinct pathogenic phenotypes in experimentally infected chickens and ferrets. J Virol. 2017;91:e01300–17. 10.1128/JVI.01300-17 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

