ATAD5 promotes replication restart by regulating RAD51 and PCNA in response to replication stress
- PMID: 31844045
- PMCID: PMC6914801
- DOI: 10.1038/s41467-019-13667-4
ATAD5 promotes replication restart by regulating RAD51 and PCNA in response to replication stress
Abstract
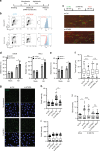
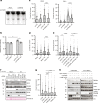
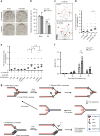
Maintaining stability of replication forks is important for genomic integrity. However, it is not clear how replisome proteins contribute to fork stability under replication stress. Here, we report that ATAD5, a PCNA unloader, plays multiple functions at stalled forks including promoting its restart. ATAD5 depletion increases genomic instability upon hydroxyurea treatment in cultured cells and mice. ATAD5 recruits RAD51 to stalled forks in an ATR kinase-dependent manner by hydroxyurea-enhanced protein-protein interactions and timely removes PCNA from stalled forks for RAD51 recruitment. Consistent with the role of RAD51 in fork regression, ATAD5 depletion inhibits slowdown of fork progression and native 5-bromo-2'-deoxyuridine signal induced by hydroxyurea. Single-molecule FRET showed that PCNA itself acts as a mechanical barrier to fork regression. Consequently, DNA breaks required for fork restart are reduced by ATAD5 depletion. Collectively, our results suggest an important role of ATAD5 in maintaining genome integrity during replication stress.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
ATAD5 restricts R-loop formation through PCNA unloading and RNA helicase maintenance at the replication fork.Nucleic Acids Res. 2020 Jul 27;48(13):7218-7238. doi: 10.1093/nar/gkaa501. Nucleic Acids Res. 2020. PMID: 32542338 Free PMC article.
-
BAP1 promotes stalled fork restart and cell survival via INO80 in response to replication stress.Biochem J. 2019 Oct 30;476(20):3053-3066. doi: 10.1042/BCJ20190622. Biochem J. 2019. PMID: 31657441
-
Deletion of BRCA2 exon 27 causes defects in response to both stalled and collapsed replication forks.Mutat Res. 2014 Aug-Sep;766-767:66-72. doi: 10.1016/j.mrfmmm.2014.06.003. Epub 2014 Jun 22. Mutat Res. 2014. PMID: 25847274
-
Is PCNA unloading the central function of the Elg1/ATAD5 replication factor C-like complex?Cell Cycle. 2013 Aug 15;12(16):2570-9. doi: 10.4161/cc.25626. Epub 2013 Jul 10. Cell Cycle. 2013. PMID: 23907118 Free PMC article. Review.
-
PCNA cycling dynamics during DNA replication and repair in mammals.Trends Genet. 2024 Jun;40(6):526-539. doi: 10.1016/j.tig.2024.02.006. Epub 2024 Mar 13. Trends Genet. 2024. PMID: 38485608 Review.
Cited by
-
Distinct Motifs in ATAD5 C-Terminal Domain Modulate PCNA Unloading Process.Cells. 2022 Jun 3;11(11):1832. doi: 10.3390/cells11111832. Cells. 2022. PMID: 35681528 Free PMC article.
-
NSMF promotes the replication stress-induced DNA damage response for genome maintenance.Nucleic Acids Res. 2021 Jun 4;49(10):5605-5622. doi: 10.1093/nar/gkab311. Nucleic Acids Res. 2021. PMID: 33963872 Free PMC article.
-
Oncogenic Kinase Cascades Induce Molecular Mechanisms That Protect Leukemic Cell Models from Lethal Effects of De Novo dNTP Synthesis Inhibition.Cancers (Basel). 2021 Jul 10;13(14):3464. doi: 10.3390/cancers13143464. Cancers (Basel). 2021. PMID: 34298678 Free PMC article.
-
Polyubiquitinated PCNA triggers SLX4-mediated break-induced replication in alternative lengthening of telomeres (ALT) cancer cells.Nucleic Acids Res. 2024 Oct 28;52(19):11785-11805. doi: 10.1093/nar/gkae785. Nucleic Acids Res. 2024. PMID: 39291733 Free PMC article.
-
ATAD5 restricts R-loop formation through PCNA unloading and RNA helicase maintenance at the replication fork.Nucleic Acids Res. 2020 Jul 27;48(13):7218-7238. doi: 10.1093/nar/gkaa501. Nucleic Acids Res. 2020. PMID: 32542338 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

