Preoperative plasma fatty acid metabolites inform risk of prostate cancer progression and may be used for personalized patient stratification
- PMID: 31842810
- PMCID: PMC6916032
- DOI: 10.1186/s12885-019-6418-2
Preoperative plasma fatty acid metabolites inform risk of prostate cancer progression and may be used for personalized patient stratification
Abstract
Background: Little is known about the relationship between the metabolite profile of plasma from pre-operative prostate cancer (PCa) patients and the risk of PCa progression. In this study we investigated the association between pre-operative plasma metabolites and risk of biochemical-, local- and metastatic-recurrence, with the aim of improving patient stratification.
Methods: We conducted a case-control study within a cohort of PCa patients recruited between 1996 and 2015. The age-matched primary cases (n = 33) were stratified in low risk, high risk without progression and high risk with progression as defined by the National Comprehensive Cancer Network. These samples were compared to metastatic (n = 9) and healthy controls (n = 10). The pre-operative plasma from primary cases and the plasma from metastatic patients and controls were assessed with untargeted metabolomics by LC-MS. The association between risk of progression and metabolite abundance was calculated using multivariate Cox proportional-hazard regression and the relationship between metabolites and outcome was calculated using median cut-off normalized values of metabolite abundance by Log-Rank test using the Kaplan Meier method.
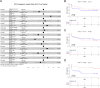
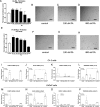
Results: Medium-chain acylcarnitines (C6-C12) were positively associated with the risk of PSA progression (p = 0.036, median cut-off) while long-chain acylcarnitines (C14-C16) were inversely associated with local (p = 0.034) and bone progression (p = 0.0033). In primary cases, medium-chain acylcarnitines were positively associated with suberic acid, which also correlated with the risk of PSA progression (p = 0.032, Log-Rank test). In the metastatic samples, this effect was consistent for hexanoylcarnitine, L.octanoylcarnitine and decanoylcarnitine. Medium-chain acylcarnitines and suberic acid displayed the same inverse association with tryptophan, while indoleacetic acid, a breakdown product of tryptophan metabolism was strongly associated with PSA (p = 0.0081, Log-Rank test) and lymph node progression (p = 0.025, Log-Rank test). These data were consistent with the increased expression of indoleamine 2,3 dioxygenase (IDO1) in metastatic versus primary samples (p = 0.014). Finally, functional experiments revealed a synergistic effect of long chain fatty acids in combination with dihydrotestosterone administration on the transcription of androgen responsive genes.
Conclusions: This study strengthens the emerging link between fatty acid metabolism and PCa progression and suggests that measuring levels of medium- and long-chain acylcarnitines in pre-operative patient plasma may provide a basis for improving patient stratification.
Keywords: Acylcarnitines; Disease progression; Fatty acid metabolism; Metabolomics; Prostate cancer.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





Similar articles
-
Identification of prediagnostic metabolites associated with prostate cancer risk by untargeted mass spectrometry-based metabolomics: A case-control study nested in the Northern Sweden Health and Disease Study.Int J Cancer. 2022 Dec 15;151(12):2115-2127. doi: 10.1002/ijc.34223. Epub 2022 Aug 12. Int J Cancer. 2022. PMID: 35866293 Free PMC article.
-
Lower Concentrations of Circulating Medium and Long-Chain Acylcarnitines Characterize Insulin Resistance in Persons with HIV.AIDS Res Hum Retroviruses. 2018 Jun;34(6):536-543. doi: 10.1089/AID.2017.0314. Epub 2018 May 2. AIDS Res Hum Retroviruses. 2018. PMID: 29607651 Free PMC article.
-
Orange juice affects acylcarnitine metabolism in healthy volunteers as revealed by a mass-spectrometry based metabolomics approach.Food Res Int. 2018 May;107:346-352. doi: 10.1016/j.foodres.2018.02.046. Epub 2018 Feb 21. Food Res Int. 2018. PMID: 29580494
-
Translational Metabolomics of Head Injury: Exploring Dysfunctional Cerebral Metabolism with Ex Vivo NMR Spectroscopy-Based Metabolite Quantification.In: Kobeissy FH, editor. Brain Neurotrauma: Molecular, Neuropsychological, and Rehabilitation Aspects. Boca Raton (FL): CRC Press/Taylor & Francis; 2015. Chapter 25. In: Kobeissy FH, editor. Brain Neurotrauma: Molecular, Neuropsychological, and Rehabilitation Aspects. Boca Raton (FL): CRC Press/Taylor & Francis; 2015. Chapter 25. PMID: 26269925 Free Books & Documents. Review.
-
Function, Detection and Alteration of Acylcarnitine Metabolism in Hepatocellular Carcinoma.Metabolites. 2019 Feb 21;9(2):36. doi: 10.3390/metabo9020036. Metabolites. 2019. PMID: 30795537 Free PMC article. Review.
Cited by
-
Palmitic acid-activated GPRs/KLF7/CCL2 pathway is involved in the crosstalk between bone marrow adipocytes and prostate cancer.BMC Cancer. 2024 Jan 15;24(1):75. doi: 10.1186/s12885-024-11826-5. BMC Cancer. 2024. PMID: 38221626 Free PMC article.
-
Targeted Metabolic Profiling in Determining the Metabolic Heterogeneity in Human Biopsies of Different Grades of Glioma.Mol Neurobiol. 2024 Oct 24. doi: 10.1007/s12035-024-04538-1. Online ahead of print. Mol Neurobiol. 2024. PMID: 39446218
-
Differential but Concerted Expression of HSD17B2, HSD17B3, SHBG and SRD5A1 Testosterone Tetrad Modulate Therapy Response and Susceptibility to Disease Relapse in Patients with Prostate Cancer.Cancers (Basel). 2021 Jul 12;13(14):3478. doi: 10.3390/cancers13143478. Cancers (Basel). 2021. PMID: 34298692 Free PMC article.
-
Plasma metabolomics of immune-related adverse events for patients with lung cancer treated with PD-1/PD-L1 inhibitors.J Immunother Cancer. 2024 Jul 11;12(7):e009399. doi: 10.1136/jitc-2024-009399. J Immunother Cancer. 2024. PMID: 38991728 Free PMC article.
-
Metabolomics based plasma biomarkers for diagnosis of oral squamous cell carcinoma and oral erosive lichen planus.J Cancer. 2022 Jan 1;13(1):76-87. doi: 10.7150/jca.59777. eCollection 2022. J Cancer. 2022. PMID: 34976172 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

