Mouse APOBEC3 interferes with autocatalytic cleavage of murine leukemia virus Pr180gag-pol precursor and inhibits Pr65gag processing
- PMID: 31830125
- PMCID: PMC6907756
- DOI: 10.1371/journal.ppat.1008173
Mouse APOBEC3 interferes with autocatalytic cleavage of murine leukemia virus Pr180gag-pol precursor and inhibits Pr65gag processing
Abstract
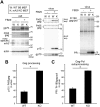
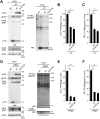
Mouse APOBEC3 (mA3) inhibits murine leukemia virus (MuLV) replication by a deamination-independent mechanism in which the reverse transcription is considered the main target process. However, other steps in virus replication that can be targeted by mA3 have not been examined. We have investigated the possible effect of mA3 on MuLV protease-mediated processes and found that mA3 binds both mature viral protease and Pr180gag-pol precursor polyprotein. Using replication-competent MuLVs, we also show that mA3 inhibits the processing of Pr65 Gag precursor. Furthermore, we demonstrate that the autoprocessing of Pr180gag-pol is impeded by mA3, resulting in reduced production of mature viral protease. This reduction appears to link with the above inefficient Pr65gag processing in the presence of mA3. Two major isoforms of mA3, exon 5-containing and -lacking ones, equally exhibit this antiviral activity. Importantly, physiologically expressed levels of mA3 impedes both Pr180gag-pol autocatalysis and Pr65gag processing. This blockade is independent of the deaminase activity and requires the C-terminal region of mA3. These results suggest that the above impairment of Pr180gag-pol autoprocessing may significantly contribute to the deaminase-independent antiretroviral activity exerted by mA3.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





Similar articles
-
Mouse APOBEC3 Restriction of Retroviruses.Viruses. 2020 Oct 27;12(11):1217. doi: 10.3390/v12111217. Viruses. 2020. PMID: 33121095 Free PMC article. Review.
-
Studies on the restriction of murine leukemia viruses by mouse APOBEC3.PLoS One. 2012;7(5):e38190. doi: 10.1371/journal.pone.0038190. Epub 2012 May 29. PLoS One. 2012. PMID: 22666481 Free PMC article.
-
Full-Length Glycosylated Gag of Murine Leukemia Virus Can Associate with the Viral Envelope as a Type I Integral Membrane Protein.J Virol. 2018 Feb 26;92(6):e01530-17. doi: 10.1128/JVI.01530-17. Print 2018 Mar 15. J Virol. 2018. PMID: 29298890 Free PMC article.
-
Enhanced replication and pathogenesis of Moloney murine leukemia virus in mice defective in the murine APOBEC3 gene.Virology. 2009 Mar 15;385(2):455-63. doi: 10.1016/j.virol.2008.11.051. Epub 2009 Jan 15. Virology. 2009. PMID: 19150103 Free PMC article.
-
Murine retrovirus escapes from murine APOBEC3 via two distinct novel mechanisms.Curr Biol. 2006 Aug 8;16(15):1565-70. doi: 10.1016/j.cub.2006.06.055. Curr Biol. 2006. PMID: 16890533
Cited by
-
Mouse APOBEC3 Restriction of Retroviruses.Viruses. 2020 Oct 27;12(11):1217. doi: 10.3390/v12111217. Viruses. 2020. PMID: 33121095 Free PMC article. Review.
-
Deaminase-Independent Mode of Antiretroviral Action in Human and Mouse APOBEC3 Proteins.Microorganisms. 2020 Dec 12;8(12):1976. doi: 10.3390/microorganisms8121976. Microorganisms. 2020. PMID: 33322756 Free PMC article. Review.
-
Retroviral Restriction Factors and Their Viral Targets: Restriction Strategies and Evolutionary Adaptations.Microorganisms. 2020 Dec 11;8(12):1965. doi: 10.3390/microorganisms8121965. Microorganisms. 2020. PMID: 33322320 Free PMC article. Review.
-
Distinctive High Expression of Antiretroviral APOBEC3 Protein in Mouse Germinal Center B Cells.Viruses. 2022 Apr 17;14(4):832. doi: 10.3390/v14040832. Viruses. 2022. PMID: 35458563 Free PMC article.
-
Myelodysplasia after clonal hematopoiesis with APOBEC3-mediated CYBB inactivation in retroviral gene therapy for X-CGD.Mol Ther. 2023 Dec 6;31(12):3424-3440. doi: 10.1016/j.ymthe.2023.09.004. Epub 2023 Sep 13. Mol Ther. 2023. PMID: 37705244 Free PMC article.
References
-
- Suspene R, Aynaud MM, Koch S, Pasdeloup D, Labetoulle M, Gaertner B, et al. Genetic editing of herpes simplex virus 1 and Epstein-Barr herpesvirus genomes by human APOBEC3 cytidine deaminases in culture and in vivo. J Virol. 2011;85(15):7594–602. Epub 2011/06/03. 10.1128/JVI.00290-11 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

