Enhancers as regulators of antigen receptor loci three-dimensional chromatin structure
- PMID: 31829768
- PMCID: PMC7053977
- DOI: 10.1080/21541264.2019.1699383
Enhancers as regulators of antigen receptor loci three-dimensional chromatin structure
Abstract
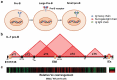
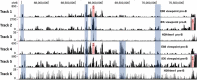
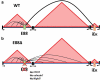
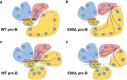
Enhancers are defined as regulatory elements that control transcription in a cell-type and developmental stage-specific manner. They achieve this by physically interacting with their cognate gene promoters. Significantly, these interactions can occur through long genomic distances since enhancers may not be near their cognate promoters. The optimal coordination of enhancer-regulated transcription is essential for the function and identity of the cell. Although great efforts to fully understand the principles of this type of regulation are ongoing, other potential functions of the long-range chromatin interactions (LRCIs) involving enhancers are largely unexplored. We recently uncovered a new role for enhancer elements in determining the three-dimensional (3D) structure of the immunoglobulin kappa (Igκ) light chain receptor locus suggesting a structural function for these DNA elements. This enhancer-mediated locus configuration shapes the resulting Igκ repertoire. We also propose a role for enhancers as critical components of sub-topologically associating domain (subTAD) formation and nuclear spatial localization.
Keywords: 3D structure; Enhancer; Igκ; V(D)J recombination; antibody repertoire; antigen receptor loci; chromatin conformation capture; long-range chromatin interactions; loop extrusion; nuclear topology; subTAD.
Figures

Similar articles
-
A B-Cell-Specific Enhancer Orchestrates Nuclear Architecture to Generate a Diverse Antigen Receptor Repertoire.Mol Cell. 2019 Jan 3;73(1):48-60.e5. doi: 10.1016/j.molcel.2018.10.013. Epub 2018 Nov 15. Mol Cell. 2019. PMID: 30449725 Free PMC article.
-
Pre-B cell receptor signaling induces immunoglobulin κ locus accessibility by functional redistribution of enhancer-mediated chromatin interactions.PLoS Biol. 2014 Feb 18;12(2):e1001791. doi: 10.1371/journal.pbio.1001791. eCollection 2014 Feb. PLoS Biol. 2014. PMID: 24558349 Free PMC article.
-
Dynamic Control of Long-Range Genomic Interactions at the Immunoglobulin κ Light-Chain Locus.Adv Immunol. 2015;128:183-271. doi: 10.1016/bs.ai.2015.07.004. Epub 2015 Aug 14. Adv Immunol. 2015. PMID: 26477368 Review.
-
Chromatin topology and the regulation of antigen receptor assembly.Annu Rev Immunol. 2012;30:337-56. doi: 10.1146/annurev-immunol-020711-075003. Epub 2012 Jan 3. Annu Rev Immunol. 2012. PMID: 22224771 Review.
-
E2A and IRF-4/Pip promote chromatin modification and transcription of the immunoglobulin kappa locus in pre-B cells.Mol Cell Biol. 2006 Feb;26(3):810-21. doi: 10.1128/MCB.26.3.810-821.2006. Mol Cell Biol. 2006. PMID: 16428437 Free PMC article.
Cited by
-
Transcriptional control by enhancers: working remotely for improved performance.Transcription. 2020 Feb;11(1):1-2. doi: 10.1080/21541264.2020.1724673. Transcription. 2020. PMID: 32054432 Free PMC article. No abstract available.
-
A Novel Framework for Characterizing Genomic Haplotype Diversity in the Human Immunoglobulin Heavy Chain Locus.Front Immunol. 2020 Sep 23;11:2136. doi: 10.3389/fimmu.2020.02136. eCollection 2020. Front Immunol. 2020. PMID: 33072076 Free PMC article.
-
Enhancers within the Ig V Gene Region Orchestrate Chromatin Topology and Regulate V Gene Rearrangement Frequency to Shape the B Cell Receptor Repertoire Specificities.J Immunol. 2023 Dec 1;211(11):1613-1622. doi: 10.4049/jimmunol.2300261. J Immunol. 2023. PMID: 37983521 Free PMC article.
-
Enhancer-instructed epigenetic landscape and chromatin compartmentalization dictate a primary antibody repertoire protective against specific bacterial pathogens.Nat Immunol. 2023 Feb;24(2):320-336. doi: 10.1038/s41590-022-01402-z. Epub 2023 Jan 30. Nat Immunol. 2023. PMID: 36717722 Free PMC article.
References
-
- Banerji J, Olson L, Schaffner W.. A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes. Cell. 1983;33(3):729–740. - PubMed
-
- Gillies SD, Morrison SL, Oi VT, et al. A tissue-specific transcription enhancer element is located in the major intron of a rearranged immunoglobulin heavy chain gene. Cell. 1983;33(3):717–728. - PubMed
-
- Queen C, Baltimore D.. Immunoglobulin gene transcription is activated by downstream sequence elements. Cell. 1983;33(3):741–748. - PubMed
-
- Schoenfelder S, Fraser P. Long-range enhancer-promoter contacts in gene expression control. Nat Rev Genet. 2019;20(8):437–455. - PubMed
-
- Sur I, Taipale J. The role of enhancers in cancer. Nat Rev Cancer. 2016;16(8):483–493. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
