Identification of Low-Abundance Lipid Droplet Proteins in Seeds and Seedlings
- PMID: 31826923
- PMCID: PMC7054876
- DOI: 10.1104/pp.19.01255
Identification of Low-Abundance Lipid Droplet Proteins in Seeds and Seedlings
Abstract
The developmental program of seed formation, germination, and early seedling growth requires not only tight regulation of cell division and metabolism, but also concerted control of the structure and function of organelles, which relies on specific changes in their protein composition. Of particular interest is the switch from heterotrophic to photoautotrophic seedling growth, for which cytoplasmic lipid droplets (LDs) play a critical role as depots for energy-rich storage lipids. Here, we present the results of a bottom-up proteomics study analyzing the total protein fractions and LD-enriched fractions in eight different developmental phases during silique (seed) development, seed germination, and seedling establishment in Arabidopsis (Arabidopsis thaliana). The quantitative analysis of the LD proteome using LD-enrichment factors led to the identification of six previously unidentified and comparably low-abundance LD proteins, each of which was confirmed by intracellular localization studies with fluorescent protein fusions. In addition to these advances in LD protein discovery and the potential insights provided to as yet unexplored aspects in plant LD functions, our data set allowed for a comparative analysis of the LD protein composition throughout the various developmental phases examined. Among the most notable of the alterations in the LD proteome were those during seedling establishment, indicating a switch in the physiological function(s) of LDs after greening of the cotyledons. This work highlights LDs as dynamic organelles with functions beyond lipid storage.
© 2020 American Society of Plant Biologists. All Rights Reserved.
Figures
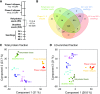
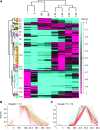

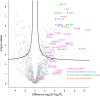
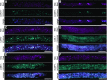


Comment in
-
Discovering Lipid Droplet Proteins: From Seeds to Seedlings.Plant Physiol. 2020 Mar;182(3):1192-1193. doi: 10.1104/pp.20.00089. Plant Physiol. 2020. PMID: 32127431 Free PMC article. No abstract available.
Similar articles
-
Lipid Droplet-Associated Proteins (LDAPs) Are Required for the Dynamic Regulation of Neutral Lipid Compartmentation in Plant Cells.Plant Physiol. 2016 Apr;170(4):2052-71. doi: 10.1104/pp.15.01977. Epub 2016 Feb 19. Plant Physiol. 2016. PMID: 26896396 Free PMC article.
-
Arabidopsis LDIP protein locates at a confined area within the lipid droplet surface and favors lipid droplet formation.Biochimie. 2020 Feb;169:29-40. doi: 10.1016/j.biochi.2019.09.018. Epub 2019 Sep 27. Biochimie. 2020. PMID: 31568826
-
PUX10 Is a Lipid Droplet-Localized Scaffold Protein That Interacts with CELL DIVISION CYCLE48 and Is Involved in the Degradation of Lipid Droplet Proteins.Plant Cell. 2018 Sep;30(9):2137-2160. doi: 10.1105/tpc.18.00276. Epub 2018 Aug 7. Plant Cell. 2018. PMID: 30087207 Free PMC article.
-
First off the mark: early seed germination.J Exp Bot. 2011 Jun;62(10):3289-309. doi: 10.1093/jxb/err030. Epub 2011 Mar 23. J Exp Bot. 2011. PMID: 21430292 Review.
-
Biogenesis and functions of lipid droplets in plants: Thematic Review Series: Lipid Droplet Synthesis and Metabolism: from Yeast to Man.J Lipid Res. 2012 Feb;53(2):215-26. doi: 10.1194/jlr.R021436. Epub 2011 Nov 1. J Lipid Res. 2012. PMID: 22045929 Free PMC article. Review.
Cited by
-
Triacylglycerol remodeling in Physaria fendleri indicates oil accumulation is dynamic and not a metabolic endpoint.Plant Physiol. 2021 Oct 5;187(2):799-815. doi: 10.1093/plphys/kiab294. Plant Physiol. 2021. PMID: 34608961 Free PMC article.
-
Heat stress leads to rapid lipid remodeling and transcriptional adaptations in Nicotiana tabacum pollen tubes.Plant Physiol. 2022 Jun 1;189(2):490-515. doi: 10.1093/plphys/kiac127. Plant Physiol. 2022. PMID: 35302599 Free PMC article.
-
Lipid droplets in Arabidopsis thaliana leaves contain myosin-binding proteins and enzymes associated with furan-containing fatty acid biosynthesis.Front Plant Sci. 2024 Mar 1;15:1331479. doi: 10.3389/fpls.2024.1331479. eCollection 2024. Front Plant Sci. 2024. PMID: 38495375 Free PMC article.
-
SEIPIN Isoforms Interact with the Membrane-Tethering Protein VAP27-1 for Lipid Droplet Formation.Plant Cell. 2020 Sep;32(9):2932-2950. doi: 10.1105/tpc.19.00771. Epub 2020 Jul 20. Plant Cell. 2020. PMID: 32690719 Free PMC article.
-
Dynamic Regulation of Lipid Droplet Biogenesis in Plant Cells and Proteins Involved in the Process.Int J Mol Sci. 2023 Apr 19;24(8):7476. doi: 10.3390/ijms24087476. Int J Mol Sci. 2023. PMID: 37108639 Free PMC article. Review.
References
-
- Baud S, Boutin JP, Miquel M, Lepiniec L, Rochat C (2002) An integrated overview of seed development in Arabidopsis thaliana ecotype WS. Plant Physiol Biochem 40: 151–160
-
- Baud S, Dichow NR, Kelemen Z, d’Andréa S, To A, Berger N, Canonge M, Kronenberger J, Viterbo D, Dubreucq B, et al. (2009) Regulation of HSD1 in seeds of Arabidopsis thaliana. Plant Cell Physiol 50: 1463–1478 - PubMed
-
- Beaudoin F, Wilkinson BM, Stirling CJ, Napier JA (2000) In vivo targeting of a sunflower oil body protein in yeast secretory (sec) mutants. Plant J 23: 159–170 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

