A 49-residue sequence motif in the C terminus of Nav1.9 regulates trafficking of the channel to the plasma membrane
- PMID: 31822564
- PMCID: PMC6983848
- DOI: 10.1074/jbc.RA119.011424
A 49-residue sequence motif in the C terminus of Nav1.9 regulates trafficking of the channel to the plasma membrane
Abstract
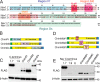
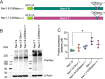
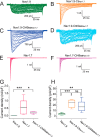
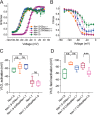
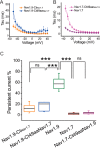
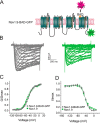
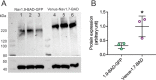
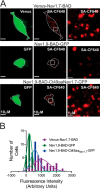
Genetic and functional studies have confirmed an important role for the voltage-gated sodium channel Nav1.9 in human pain disorders. However, low functional expression of Nav1.9 in heterologous systems (e.g. in human embryonic kidney 293 (HEK293) cells) has hampered studies of its biophysical and pharmacological properties and the development of high-throughput assays for drug development targeting this channel. The mechanistic basis for the low level of Nav1.9 currents in heterologous expression systems is not understood. Here, we implemented a multidisciplinary approach to investigate the mechanisms that govern functional Nav1.9 expression. Recombinant expression of a series of Nav1.9-Nav1.7 C-terminal chimeras in HEK293 cells identified a 49-amino-acid-long motif in the C terminus of the two channels that regulates expression levels of these chimeras. We confirmed the critical role of this motif in the context of a full-length channel chimera, Nav1.9-Ct49aaNav1.7, which displayed significantly increased current density in HEK293 cells while largely retaining the characteristic Nav1.9-gating properties. High-resolution live microscopy indicated that the newly identified C-terminal motif dramatically increases the number of channels on the plasma membrane of HEK293 cells. Molecular modeling results suggested that this motif is exposed on the cytoplasmic face of the folded C terminus, where it might interact with other channel partners. These findings reveal that a 49-residue-long motif in Nav1.9 regulates channel trafficking to the plasma membrane.
Keywords: Nav1.9; channel trafficking; electrophysiology; functional expression; live imaging; nociception; pain; sensory neurons; sodium channel; structural model; trafficking; voltage clamp.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures

Similar articles
-
Heterologous expression of NaV1.9 chimeras in various cell systems.Pflugers Arch. 2015 Dec;467(12):2423-35. doi: 10.1007/s00424-015-1709-1. Epub 2015 Apr 29. Pflugers Arch. 2015. PMID: 25916202
-
Biophysical and Pharmacological Characterization of Nav1.9 Voltage Dependent Sodium Channels Stably Expressed in HEK-293 Cells.PLoS One. 2016 Aug 24;11(8):e0161450. doi: 10.1371/journal.pone.0161450. eCollection 2016. PLoS One. 2016. PMID: 27556810 Free PMC article.
-
Spider venom-derived peptide induces hyperalgesia in Nav1.7 knockout mice by activating Nav1.9 channels.Nat Commun. 2020 May 8;11(1):2293. doi: 10.1038/s41467-020-16210-y. Nat Commun. 2020. PMID: 32385249 Free PMC article.
-
Dynamic compartmentalization of the voltage-gated sodium channels in axons.Biol Cell. 2003 Oct;95(7):437-45. doi: 10.1016/s0248-4900(03)00091-1. Biol Cell. 2003. PMID: 14597261 Review.
-
Structure-based assessment of disease-related mutations in human voltage-gated sodium channels.Protein Cell. 2017 Jun;8(6):401-438. doi: 10.1007/s13238-017-0372-z. Epub 2017 Feb 1. Protein Cell. 2017. PMID: 28150151 Free PMC article. Review.
Cited by
-
Reduced pain sensitivity of episodic pain syndrome model mice carrying a Nav1.9 mutation by ANP-230, a novel sodium channel blocker.Heliyon. 2023 Apr 14;9(4):e15423. doi: 10.1016/j.heliyon.2023.e15423. eCollection 2023 Apr. Heliyon. 2023. PMID: 37151704 Free PMC article.
-
Homozygous C-terminal loss-of-function NaV1.4 variant in a patient with congenital myasthenic syndrome.J Neurol Neurosurg Psychiatry. 2020 Aug;91(8):898-900. doi: 10.1136/jnnp-2020-323173. Epub 2020 Jun 2. J Neurol Neurosurg Psychiatry. 2020. PMID: 32487525 Free PMC article. No abstract available.
-
Pharmacological activity and NMR solution structure of the leech peptide HSTX-I.Biochem Pharmacol. 2020 Nov;181:114082. doi: 10.1016/j.bcp.2020.114082. Epub 2020 Jun 7. Biochem Pharmacol. 2020. PMID: 32524995 Free PMC article.
-
Direct fluorescent labeling of NF186 and NaV1.6 in living primary neurons using bioorthogonal click chemistry.J Cell Sci. 2023 Jun 15;136(12):jcs260600. doi: 10.1242/jcs.260600. Epub 2023 Jun 28. J Cell Sci. 2023. PMID: 37288813 Free PMC article.
-
Characterization of Synthetic Tf2 as a NaV1.3 Selective Pharmacological Probe.Biomedicines. 2020 Jun 11;8(6):155. doi: 10.3390/biomedicines8060155. Biomedicines. 2020. PMID: 32545167 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

