Cryo-EM structure of the human MLL1 core complex bound to the nucleosome
- PMID: 31804488
- PMCID: PMC6895043
- DOI: 10.1038/s41467-019-13550-2
Cryo-EM structure of the human MLL1 core complex bound to the nucleosome
Erratum in
-
Publisher Correction: Cryo-EM structure of the human MLL1 core complex bound to the nucleosome.Nat Commun. 2020 Feb 27;11(1):1165. doi: 10.1038/s41467-020-14973-y. Nat Commun. 2020. PMID: 32109228 Free PMC article.
Abstract
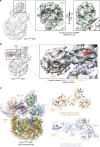
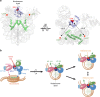
Mixed lineage leukemia (MLL) family histone methyltransferases are enzymes that deposit histone H3 Lys4 (K4) mono-/di-/tri-methylation and regulate gene expression in mammals. Despite extensive structural and biochemical studies, the molecular mechanisms whereby the MLL complexes recognize histone H3K4 within nucleosome core particles (NCPs) remain unclear. Here we report the single-particle cryo-electron microscopy (cryo-EM) structure of the NCP-bound human MLL1 core complex. We show that the MLL1 core complex anchors to the NCP via the conserved RbBP5 and ASH2L, which interact extensively with nucleosomal DNA and the surface close to the N-terminal tail of histone H4. Concurrent interactions of RbBP5 and ASH2L with the NCP uniquely align the catalytic MLL1SET domain at the nucleosome dyad, thereby facilitating symmetrical access to both H3K4 substrates within the NCP. Our study sheds light on how the MLL1 complex engages chromatin and how chromatin binding promotes MLL1 tri-methylation activity.
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
An Ash2L/RbBP5 heterodimer stimulates the MLL1 methyltransferase activity through coordinated substrate interactions with the MLL1 SET domain.PLoS One. 2010 Nov 23;5(11):e14102. doi: 10.1371/journal.pone.0014102. PLoS One. 2010. PMID: 21124902 Free PMC article.
-
Structural basis of nucleosome recognition and modification by MLL methyltransferases.Nature. 2019 Sep;573(7774):445-449. doi: 10.1038/s41586-019-1528-1. Epub 2019 Sep 4. Nature. 2019. PMID: 31485071
-
Automethylation activities within the mixed lineage leukemia-1 (MLL1) core complex reveal evidence supporting a "two-active site" model for multiple histone H3 lysine 4 methylation.J Biol Chem. 2014 Jan 10;289(2):868-84. doi: 10.1074/jbc.M113.501064. Epub 2013 Nov 14. J Biol Chem. 2014. PMID: 24235145 Free PMC article.
-
The Core Complex of Yeast COMPASS and Human Mixed-Lineage Leukemia (MLL), Structure, Function, and Recognition of the Nucleosome.Subcell Biochem. 2024;104:101-117. doi: 10.1007/978-3-031-58843-3_6. Subcell Biochem. 2024. PMID: 38963485 Review.
-
The Development of Inhibitors Targeting the Mixed Lineage Leukemia 1 (MLL1)-WD Repeat Domain 5 Protein (WDR5) Protein- Protein Interaction.Curr Med Chem. 2020;27(33):5530-5542. doi: 10.2174/0929867326666190528080514. Curr Med Chem. 2020. PMID: 31132972 Review.
Cited by
-
Parallel functional annotation of cancer-associated missense mutations in histone methyltransferases.Sci Rep. 2022 Nov 2;12(1):18487. doi: 10.1038/s41598-022-23229-2. Sci Rep. 2022. PMID: 36323913 Free PMC article.
-
Multistate structures of the MLL1-WRAD complex bound to H2B-ubiquitinated nucleosome.Proc Natl Acad Sci U S A. 2022 Sep 20;119(38):e2205691119. doi: 10.1073/pnas.2205691119. Epub 2022 Sep 12. Proc Natl Acad Sci U S A. 2022. PMID: 36095189 Free PMC article.
-
Human WDR5 promotes breast cancer growth and metastasis via KMT2-independent translation regulation.Elife. 2022 Aug 31;11:e78163. doi: 10.7554/eLife.78163. Elife. 2022. PMID: 36043466 Free PMC article.
-
The Ash2l SDI Domain Is Required to Maintain the Stability and Binding of DPY30.Cells. 2022 Apr 25;11(9):1450. doi: 10.3390/cells11091450. Cells. 2022. PMID: 35563756 Free PMC article.
-
Navigating the structure of COMPASS.Elife. 2020 Feb 24;9:e54767. doi: 10.7554/eLife.54767. Elife. 2020. PMID: 32091392 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

