TRIM21-mediated proteasomal degradation of SAMHD1 regulates its antiviral activity
- PMID: 31797533
- PMCID: PMC6944907
- DOI: 10.15252/embr.201847528
TRIM21-mediated proteasomal degradation of SAMHD1 regulates its antiviral activity
Abstract
SAMHD1 possesses multiple functions, but whether cellular factors regulate SAMHD1 expression or its function remains not well characterized. Here, by investigating why cultured RD and HEK293T cells show different sensitivity to enterovirus 71 (EV71) infection, we demonstrate that SAMHD1 is a restriction factor for EV71. Importantly, we identify TRIM21, an E3 ubiquitin ligase, as a key regulator of SAMHD1, which specifically interacts and degrades SAMHD1 through the proteasomal pathway. However, TRIM21 has no effect on EV71 replication itself. Moreover, we prove that interferon production stimulated by EV71 infection induces increased TRIM21 and SAMHD1 expression, whereas increasing TRIM21 overrides SAMHD1 inhibition of EV71 in cells and in a neonatal mouse model. TRIM21-mediated degradation of SAMHD1 also affects SAMHD1-dependent restriction of HIV-1 and the regulation of interferon production. We further identify the functional domains in TRIM21 required for SAMHD1 binding and the ubiquitination site K622 in SAMHD1 and show that phosphorylation of SAMHD1 at T592 also blocks EV71 restriction. Our findings illuminate how EV71 overcomes SAMHD1 inhibition via the upregulation of TRIM21.
Keywords: EV71 infection; SAMHD1 inhibition; interferon induction; regulation; ubiquitin-proteasome degradation.
© 2019 The Authors. Published under the terms of the CC BY 4.0 license.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
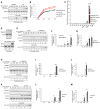
- A–D
The expression of SAMHD1 is negatively correlated with EV71 viral replication ability. HEK293T and RD cells were infected with EV71 at a MOI of 0.1; then, the cells and supernatants were harvested at the indicated time points. (A) Immunoblotting (IB) analysis of EV71 VP1 in cells, with tubulin as a loading control. EV71‐VP1 protein in the supernatants was detected after ultracentrifugation. (B) EV71 viral RNA levels in cell lysates and supernatants were detected by RT–qPCR with GAPDH as a control. (n = 3, mean ± SD, ns stands for no significance, paired t‐test) (C) The mRNA levels of the host restriction factors were detected by RT–qPCR in infected or uninfected HEK293T or RD cells, and the expression levels of the target genes were normalized to GAPDH. (n = 3, mean ± SD, ns stands for no significance, paired t‐test). (D) IB analysis of SAMHD1 and BST‐2 protein levels with tubulin as a loading control. The densities of bands were analyzed with ImageJ software to calculate the values relative to that of tubulin.
- E–J
SAMHD1 knockdown enhances EV71 replication. The stable cell lines pLKO.1 and sh‐SAMHD1 constructed in HEK293T (E) and RD (H) cells were infected with EV71 at a MOI of 0.1 and 0.05, respectively, and cells and supernatants were harvested at the indicated time points. IB analysis of EV71 VP1 and SAMHD1 in cells was performed with tubulin as a loading control. EV71‐VP1 protein in the supernatants was detected after ultracentrifugation. (F and I) EV71 viral RNA in cell lysates was detected by RT–qPCR with GAPDH as a control (n = 3, mean ± SD, *P < 0.05, **P < 0.01, paired t‐test). (G and J) Viral titers in the supernatants were measured by the cytopathic effect method. The results represent the means ± SD from three independent experiments. Statistical significance was analyzed using Student's t‐test (*P < 0.05, **P < 0.01).
- K
HEK293T cells transfected with VR1012, which is a eukaryotic expression vector, or Vpx‐HA were infected with EV71 at a MOI of 0.1, and then, cells and supernatants were harvested at the indicated time points. IB analysis of EV71‐VP1, SAMHD1, and Vpx‐HA in cell lysates was performed with tubulin as a loading control. EV71‐VP1 was detected in the supernatants after ultracentrifugation.
- L
EV71 viral RNA was detected in cell lysates by RT–qPCR with GAPDH as a control (n = 3, mean ± SD, **P < 0.01, paired t‐test).
- M
Viral titers in the supernatants were measured by the cytopathic effect method. The results represent the means ± SD from three independent experiments. Statistical significance was analyzed using Student's t‐test (**P < 0.01).
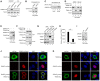
- A
Stable SAMHD1‐HA‐overexpressing cell lines were constructed in HEK293T and RD cells and detected by IB with pLVX as a negative control.
- B
RD SAMHD1‐HA cell lines were treated with dimethyl sulfoxide (DMSO) or 10 μM MG132 for 12 h prior to harvest and subjected to IB with tubulin as a loading control.
- C
RD SAMHD1‐HA cell lines were treated as in (B), and then, HA immunoprecipitation (IP) and IB were performed. The IP elution was analyzed by mass spectrometry.
- D, E
TRIM21 (D) but not TRIM25 (E) induced the degradation of SAMHD1, and MG132 rescued the TRIM21‐mediated degradation of SAMHD1. HEK293T cells were cotransfected with SAMHD1 and VR1012 or TRIM21 or TRIM25 expression plasmids with or without MG132 treatment for 12 h prior to harvest and then subjected to IB with tubulin as a loading control.
- F
Reverse co‐IP confirmed the interaction between SAMHD1 and TRIM21. SAMHD1‐flag was transfected with TRIM21‐HA into HEK293T cells, and then, the cells were treated as in (B) and subjected to HA IP and IB.
- G
Endogenous TRIM21 but not TRIM25 still interacted with SAMHD1. RD‐SAMHD1‐HA cells treated as in (B) were subjected to TRIM21 or TRIM25 IP and IB.
- H, I
(H) mRNA (n = 3, mean ± SD, ***P < 0.001, paired t‐test) and (I) protein levels of TRIM21 in RD and HEK293T cells.
- J, K
TRIM21 (J) but not TRIM25 (K) colocalized with SAMHD1. Images were taken under a Zeiss LZM710 confocal microscope, Bars, 10 μm.

IB analysis of RD cells treated with scrambled shRNA or with TRIM21‐specific shRNA with tubulin as a loading control.
RD‐pLKO.1 or RD‐shTRIM21‐3 cells were infected with EV71 at a MOI of 0.05 for the indicated time and harvested for SAMHD1, TRIM21, and EV71‐VP1 detection by IB. Tubulin served as a loading control.
EV71 RNA levels in (B) were detected by RT–qPCR with GAPDH as a control (n = 3, mean ± SD, **P < 0.01, paired t‐test).
PBMCs were isolated from three healthy donors and infected with EV71 at 0.1 MOI. After 72 h, PBMCs were harvested and subjected to IB for SAMHD1, TRIM21, and EV71‐VP1 detection with tubulin as a control.
Viral mRNA level of EV71 was detected in PBMCs by RT–qPCR with GAPDH as a control (n = 3, mean ± SD).

- A–D
TRIM21 knockout in RD cells released SAMHD1 inhibition on EV71 replication. (A) Illustration of gRNA‐guided TRIM21 knockout in cells. (B) Negative control or RD‐CRISPR‐Cas9‐TRIM21 RD cells were infected with EV71 at a MOI of 0.05 for the indicated time and harvested for SAMHD1, TRIM21, and EV71‐VP1 detection by IB. Tubulin served as a loading control. (C) EV71 RNA levels in (B) were detected by RT–qPCR with GAPDH as a control (n = 3, mean ± SD, **P < 0.01, paired t‐test). (D) Viral titers in the supernatant were measured by the cytopathic effect method. The results represent the means ± SD from three independent experiments. Statistical significance was analyzed using Student's t‐test (**P < 0.01).
- E, F
Overexpression of TRIM21 in HEK293T cells increased EV71 replication by degrading SAMHD1. HEK293T cells were transfected with VR1012 or TRIM21‐HA for 24 h and then infected with EV71 at a MOI of 0.05. (E) Cells were harvested at the indicated time points and subjected to IB with tubulin as a loading control. (F) EV71 viral RNA in cell lysates was detected by RT–qPCR with GAPDH as a control (n = 3, mean ± SD, **P < 0.01, paired t‐test).
- G, H
TRIM21 itself has no effect on EV71 replication. VR1012 or TRIM21 was transfected into pLKO.1 or sh‐SAMHD1 HEK293T cells for 24 h and infected with EV71. (G) Cells were harvested and subjected to IB analysis. (H) Viral titers in the supernatants were measured by the cytopathic effect method. The results represent the means ± SD from three independent experiments. Statistical significance was analyzed using Student's t‐test (*P < 0.05).

- A, B
Stable cell lines scrambled gRNA or TRIM21 gRNA constructed in RD cells were transfected with siRNA‐NC or siRNA‐SAMHD1, and then infected with EV71 at 0.1 MOI. After 48 h, cells and supernatants were harvested and analysis by IB (A) and titer detection (B) Viral titers in the supernatants were measured by the cytopathic effect method. The results represent the means ± SD from three independent experiments. Statistical significance was analyzed using Student's t‐test (*P < 0.05, **P < 0.01).
- C
Stable cell lines scrambled gRNA or TRIM21 gRNA constructed in RD cells were treated with 100 μg/ml cycloheximide (CHX) and DMSO or 10 μM MG132. Cells were harvested at the indicated time points and then analyzed by IB. The densities of bands were analyzed with ImageJ software to calculate the values relative to that for tubulin.
- D, E
Stable cell lines pLKO.1 or sh‐SAMHD1 constructed in HEK293T cells were infected with RSV at a MOI of 0.1, and then, the cells were harvested at the indicated time points. IB analysis of RSV proteins and SAMHD1 was performed with tubulin as a loading control. (F) SAMHD1 inhibits VSV within 24 h. VR1012 or TRIM21 was transfected into pLKO.1 or sh‐SAMHD1 HEK293T cells for 24 h and then infected with VSV‐GFP at 0.01 MOI. At the indicated time points, the infectivity of VSV was measured by enumerating GFP‐positive cells. (n = 3, mean ± SD, *P < 0.05, **P < 0.01, paired t‐test).
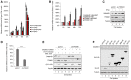
- A, B
mRNA level of IFN‐α and IFN‐β as well as SAMHD1 and TRIM21 was upregulated upon EV71 infection. HEK293T and RD cells were infected with EV71 at 0.05 MOI and harvested at different time, and then, mRNA level of IFN‐α, IFN‐β (A), TRIM21, and SAMHD1 (B) was detected by RT–qPCR with GAPDH as a control (n = 3, mean ± SD).
- C
TRIM21 is IFN‐induced protein. THP1‐pLKO.1 or THP1‐shTRIM21 cells were stimulated with IFN‐α (100 U/ml) for 24 h, and then, SAMHD1 and TRIM21 were detected by Western blot with tubulin as a control.
- D, E
Upregulation of TRIM21 by EV71 infection is IFNAR1‐dependent. (D) IFNAR knockdown was confirmed by RT–qPCR detection (n = 3, mean ± SD, ***P < 0.001, paired t‐test). (E) RD‐pLKO.1 and RD‐shIFNAR cells were transfected with SAMHD1 siRNA as indicated, then infected with EV71 at 0.05 MOI, and harvested for SAMHD1, TRIM21, and EV71‐VP1 detection 72 h post‐infection by IB with tubulin as a control.
- F
EV71 nonstructural proteins have no effect on SAMHD1 expression. HEK293T cells were transfected with SAMHD1 plus VR1012 or the indicated EV71 nonstructural proteins for 48 h and subjected to IB analysis with tubulin as a loading control; the blots of EV71 nonstructural proteins were asterisked.
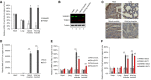
- A
The mRNA levels of SAMHD1 and TRIM21 in different tissues of neonatal mice were detected by RT–qPCR with GAPDH as a control, and the expression of SAMHD1 and TRIM21 in the heart was set as 100% (n = 3, mean ± SD, *P < 0.05, paired t‐test).
- B
The protein levels of SAMHD1 and TRIM21 in different tissues of mice were examined by near‐infrared spectrum (NIR) Western blot with tubulin as a control.
- C
Immunohistochemical (IHC) staining results of SAMHD1 in various tissues of mice. Bars, 100 μm.
- D–F
The mRNA levels of EV71 virus and host factors in mock‐ or EV71‐infected mice were detected by RT–qPCR with GAPDH as a control. The expression of EV71 (D), IFN‐α and IFN‐β (E), and SAMHD1 and TRIM21 (F) were presented, and the levels of corresponding gene in mock‐infected mice were set as 100% (n = 3, mean ± SD, **P < 0.01, paired t‐test).

- A–I
(A) IB analysis of THP‐1 cells treated with scrambled shRNA, TRIM21‐specific shRNA, or SAMHD1‐specific shRNA. Tubulin served as loading control. (B, E, H) Quantification of SAMHD1 and TRIM21 levels in whole cells in which the indicated genes were knocked down by shRNA. Data are normalized to tubulin and expressed as the fold change over cells treated with pLKO.1 without HIV infection (n = 3, mean ± SD). (C) sh‐SAMHD1 and sh‐TRIM21 THP‐1 cells were treated with PMA for 24 h and infected with the VSV‐G pseudotyped reporter HIV‐1, and then detected 48 h later by flow cytometry analysis (n = 3, mean ± SD, *P < 0.05, **P < 0.01, ***P < 0.001, paired t‐test). (D) IB analysis of MDM cells from three donors transfected with TRIM21 siRNA twice at days 1 and 3, respectively, after MDM differentiation. (F) MDM cells treated with TRIM21 siRNA were infected with VSV‐G‐pseudotyped HIV‐1 reporter and then detected 48 h later by flow cytometry analysis (n = 3, mean ± SD, **P < 0.01, paired t‐test). (G) IB analysis of Jurkat and KG‐1 cells treated with scrambled shRNA or TRIM21‐specific shRNA. (I) Jurkat and KG‐1 cells treated with TRIM21 shRNA were infected with HIV‐1 and then detected 48 h later by flow cytometry analysis (n = 3, mean ± SD, ns stands for no significance, paired t‐test).
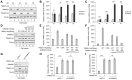
- A–C
TRIM21 enhanced the expression of IFN‐α and IFN‐β. TRIM21 was transfected into HEK293T cells for 48 h and then treated with hydroxyurea (HU) for the indicated time points. (A) IB analysis of SAMHD1. mRNA levels of IFN‐α (B) and IFN‐β (C) after treatment with HU (n = 3, mean ± SD, *P < 0.05, **P < 0.01, paired t‐test).
- D–F
Increasing SAMHD1 reduced the TRIM21‐induced increase in of IFN‐α and IFN‐β mRNA. HEK293T cells were cotransfected with TRIM21 WT or C16A mutant and increasing doses of SAMHD1 for 48 h and then treated with HU for 4 h. Cells were subjected to IB analysis (D), and the levels of IFN‐α (E) and IFN‐β (F) mRNA were detected by RT–qPCR with GAPDH as a control (n = 3, mean ± SD).
- G–I
TRIM21 enhanced IFN‐α and IFN‐β production in a SAMHD1‐dependent manner. TRIM21 was transfected into pLKO.1 or sh‐SAMHD1 HEK293T cells for 48 h, and then, the cells were treated with HU for 4 h. Cells were subjected to IB analysis (G), and the levels of IFN‐α (H) and IFN‐β (I) mRNA were detected by RT–qPCR with GAPDH as a control (n = 3, mean ± SD, **P < 0.01, paired t‐test).
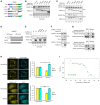
- A–C
TRIM21 interacts with SAMHD1 via PRY and SPRY domains. (A) Sketch map of TRIM21 WT and mutants. (B) The effect of TRIM21 on the degradation of SAMHD1. SAMHD1‐flag was cotransfected with VR1012, TRIM21 WT, or the indicated mutant into HEK293T cells for 48 h, and the cells were subjected to IB with tubulin as loading control. (C) SAMHD1‐flag was cotransfected with VR1012 or TRIM21 WT or the indicated mutant for 24 h, and the cells were then treated with 10 μM MG132 for 12 h before harvest and subjected to HA IP and IB.
- D
Map of SAMHD1 WT and truncation mutants.
- E
The effect of TRIM21 on SAMHD1 WT or its mutants. SAMHD1‐HA or the indicated mutant were cotransfected with VR1012 or TRIM21‐flag into HEK293T cells for 48 h, and the cells were subjected to IB with tubulin as a loading control.
- F
SAMHD1‐flag 1–547 was cotransfected with VR1012 or TRIM21‐HA for 24 h, and the cells were then treated with 10 μM MG132 for 12 h before harvest and subjected to HA IP and IB.
- G
SAMHD1 109–626 followed with a His tag or TRIM21‐PRYSPRY followed with a GST tag was expressed in Rosetta (DE3), and pull‐down assay was performed with Ni Sepharose (up) and GST Sepharose (down), respectively.
- H
FRET analysis indicates interaction between YFP‐SAMHD1 and CFP‐TRIM21. A representative image of SAMHD1‐YFP (yellow) and ECFP‐TRIM21 (cyan)‐expressing cells before and after photobleaching the acceptor fluorophore, YFP. The region chosen for photobleaching is marked (white open box), Bars, 10 μm. The quantization of fluorescence brightness was analyzed by ImageJ (n = 3, mean ± SD, ns stands for no significance, *P < 0.05, paired t‐test).
- I
The microscale thermophoresis curve for the interaction between SAMHD1 and TRIM21. Alexa Fluor® 647‐labeled SAMHD1 at a concentration of 20 nM was incubated with twofold dilution series of unlabeled TRIM21 (3 μM to 9.16E‐05 μM). The curve represents the signal recorded from three measurements. The normalized fluorescence thermophoretic signals were plotted against the concentration of TRIM21 with mean values ± standard deviation as well as a fitting to a 1:1 binding model (NanoTemper® Analysis software, F Norm = FHot/FCold). The K d of this interaction was determined to be 270 ± 92 nM. The data were a representative of three independent experiments using different dilution.

The effect of TRIM21 on SAMHD1 proteins from various species. HEK293T cells were transfected with VR1012 or TRIM21 and the indicated SAMHD1 expression vector and then subjected to IB analysis.
Identification of amino acids presented only in SAMHD1 of Canis and Gallus but not in other SAMHD1 proteins.
Construction of hSAMHD1 mutants with amino acid alterations.
The effect of TRIM21 on hSAMHD1 mutants. HEK293T cells were transfected with VR1012 or TRIM21 and SAMHD1 mutants for 48 h and then subjected to IB analysis.
SAMHD1‐flag WT or the G153S or G183R mutant was cotransfected with VR1012 or TRIM21‐HA for 24 h, and the cells were then treated with 10 μM MG132 for 12 h before harvest and subjected to HA IP and IB.
Sketch map of potential ubiquitination sites on SAMHD1.
SAMHD1‐HA WT or the indicated mutants were cotransfected with VR1012 or TRIM21‐flag into HEK293T cells for 48 h, and the cells were subjected to IB with tubulin as a loading control.
SAMHD1‐HA WT and VR1012 or TRIM21 were cotransfected with K48‐only and K63‐only ubiquitin‐flag into HEK293T cells for 24 h, and the cells were then treated with 10 μM MG132 for 12 h before harvest and subjected to HA IP and IB with tubulin as a loading control.
K48‐only ubiquitin‐flag and SAMHD1‐HA WT or mutant K622R were cotransfected with VR1012 or TRIM21 into HEK293T cells as indicated. The cells were then treated with 10 μM MG132 for 12 h before harvest and subjected to HA IP and IB with tubulin as a loading control.
VR1012, SAMHD1‐HA WT, or the K622R mutant was transfected into HEK293T‐shSAMHD1 cells for 24 h. The cells were then infected with EV71 at a MOI of 0.1 and harvested at the indicated time points. SAMHD1‐HA, TRIM21, and EV71‐VP1 were detected by IB with tubulin as a loading control.

- A
Stable sh‐SAMHD1 HEK293T cells were transfected with VR1012, SAMHD1 WT, or the indicated mutant for 24 h and then infected with EV71 at a MOI of 0.05 for 72 h. Cells were harvested and subjected to IB analysis.
- B
IB analysis of T592‐phosphorylated SAMHD1 and total SAMHD1 in RD and HEK293T cells.
- C
The effect of TRIM21 on endogenous phosphorylated SAMHD1 and total SAMHD1.
- D
The effect of TRIM21 on ectopic phosphorylated and unphosphorylated SAMHD1.
- E, F
TRIM21 directly induced the degradation of SAMHD1 but not via changing cell cycle or upregulating cyclin L2 expression. (E) TRIM21 has no effect on the G0–G1 transition in MDM. (F) TRIM21 has no effect on the expression of cyclin L2 in MDM.
Similar articles
-
A Cyclin-Binding Motif in Human SAMHD1 Is Required for Its HIV-1 Restriction, dNTPase Activity, Tetramer Formation, and Efficient Phosphorylation.J Virol. 2018 Feb 26;92(6):e01787-17. doi: 10.1128/JVI.01787-17. Print 2018 Mar 15. J Virol. 2018. PMID: 29321329 Free PMC article.
-
SAMHD1 Inhibits Multiple Enteroviruses by Interfering with the Interaction between VP1 and VP2 Proteins.J Virol. 2021 Jun 10;95(13):e0062021. doi: 10.1128/JVI.00620-21. Epub 2021 Jun 10. J Virol. 2021. PMID: 33883225 Free PMC article.
-
Inhibition of Vpx-Mediated SAMHD1 and Vpr-Mediated Host Helicase Transcription Factor Degradation by Selective Disruption of Viral CRL4 (DCAF1) E3 Ubiquitin Ligase Assembly.J Virol. 2017 Apr 13;91(9):e00225-17. doi: 10.1128/JVI.00225-17. Print 2017 May 1. J Virol. 2017. PMID: 28202763 Free PMC article.
-
[Research Progress in Viral Protein Vpx induction of Proteasomal Degradation of the Antiviral Factor SAMHD1].Bing Du Xue Bao. 2016 May;32(3):355-60. Bing Du Xue Bao. 2016. PMID: 29963825 Review. Chinese.
-
Roles of SAMHD1 in antiviral defense, autoimmunity and cancer.Rev Med Virol. 2017 Jul;27(4). doi: 10.1002/rmv.1931. Epub 2017 Apr 25. Rev Med Virol. 2017. PMID: 28444859 Review.
Cited by
-
Functional skewing of TRIM21-SIRT5 interplay dictates IL-1β production in DSS-induced colitis.EMBO Rep. 2022 Sep 5;23(9):e54391. doi: 10.15252/embr.202154391. Epub 2022 Jun 30. EMBO Rep. 2022. PMID: 35770730 Free PMC article.
-
TRIM21 inhibits porcine epidemic diarrhea virus proliferation by proteasomal degradation of the nucleocapsid protein.Arch Virol. 2021 Jul;166(7):1903-1911. doi: 10.1007/s00705-021-05080-4. Epub 2021 Apr 26. Arch Virol. 2021. PMID: 33900472 Free PMC article.
-
SAMHD1 silencing cooperates with radiotherapy to enhance anti-tumor immunity through IFI16-STING pathway in lung adenocarcinoma.J Transl Med. 2022 Dec 29;20(1):628. doi: 10.1186/s12967-022-03844-3. J Transl Med. 2022. PMID: 36578072 Free PMC article.
-
Impaired influenza A virus replication by the host restriction factor SAMHD1 which inhibited by PA-mediated dephosphorylation of the host transcription factor IRF3.Virol J. 2024 Jan 29;21(1):33. doi: 10.1186/s12985-024-02295-0. Virol J. 2024. PMID: 38287375 Free PMC article.
-
PIAS1 potentiates the anti-EBV activity of SAMHD1 through SUMOylation.Cell Biosci. 2021 Jul 8;11(1):127. doi: 10.1186/s13578-021-00636-y. Cell Biosci. 2021. PMID: 34238351 Free PMC article.
References
-
- Li N, Zhang W, Cao X (2000) Identification of human homologue of mouse IFN‐gamma induced protein from human dendritic cells. Immunol Lett 74: 221–224 - PubMed
-
- Chen S, Bonifati S, Qin Z, St Gelais C, Kodigepalli KM, Barrett BS, Kim SH, Antonucci JM, Ladner KJ, Buzovetsky O et al (2018) SAMHD1 suppresses innate immune responses to viral infections and inflammatory stimuli by inhibiting the NF‐kappaB and interferon pathways. Proc Natl Acad Sci USA 115: E3798–E3807 - PMC - PubMed
-
- Coquel F, Silva MJ, Techer H, Zadorozhny K, Sharma S, Nieminuszczy J, Mettling C, Dardillac E, Barthe A, Schmitz AL et al (2018) SAMHD1 acts at stalled replication forks to prevent interferon induction. Nature 557: 57–61 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 81672004/NSF | Foundation for Innovative Research Groups of the National Natural Science Foundation of China/International
- 31270202/NSF | Foundation for Innovative Research Groups of the National Natural Science Foundation of China/International
- 81701987/NSF | Foundation for Innovative Research Groups of the National Natural Science Foundation of China/International
- JLUSTIRT, 2017TD-05/JLU | Program for Jilin University Science and Technology Innovative Research Team (Program for JLU Science and Technology Innovative Research Team)/International
- 20190101003JH/Science and Technology Department of Jilin Province/International
- 20102209/Key Laboratory of Molecular Virology, Jilin Province/International
- JDYY82017003/Youth Foundation of the First Hospital of Jilin University/International
- Graduate Innovation Fund of Jilin University/International
- 2018ZX10302104-001-010/Chinese Ministry of Science and Technology/International
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

