Alu insertion polymorphisms shared by Papio baboons and Theropithecus gelada reveal an intertwined common ancestry
- PMID: 31788036
- PMCID: PMC6880559
- DOI: 10.1186/s13100-019-0187-y
Alu insertion polymorphisms shared by Papio baboons and Theropithecus gelada reveal an intertwined common ancestry
Abstract
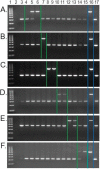
Background: Baboons (genus Papio) and geladas (Theropithecus gelada) are now generally recognized as close phylogenetic relatives, though morphologically quite distinct and generally classified in separate genera. Primate specific Alu retrotransposons are well-established genomic markers for the study of phylogenetic and population genetic relationships. We previously reported a computational reconstruction of Papio phylogeny using large-scale whole genome sequence (WGS) analysis of Alu insertion polymorphisms. Recently, high coverage WGS was generated for Theropithecus gelada. The objective of this study was to apply the high-throughput "poly-Detect" method to computationally determine the number of Alu insertion polymorphisms shared by T. gelada and Papio, and vice versa, by each individual Papio species and T. gelada. Secondly, we performed locus-specific polymerase chain reaction (PCR) assays on a diverse DNA panel to complement the computational data.
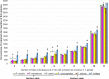
Results: We identified 27,700 Alu insertions from T. gelada WGS that were also present among six Papio species, with nearly half (12,956) remaining unfixed among 12 Papio individuals. Similarly, each of the six Papio species had species-indicative Alu insertions that were also present in T. gelada. In general, P. kindae shared more insertion polymorphisms with T. gelada than did any of the other five Papio species. PCR-based genotype data provided additional support for the computational findings.
Conclusions: Our discovery that several thousand Alu insertion polymorphisms are shared by T. gelada and Papio baboons suggests a much more permeable reproductive barrier between the two genera then previously suspected. Their intertwined evolution likely involves a long history of admixture, gene flow and incomplete lineage sorting.
Keywords: Alu element; Evolutionary biology; Primate phylogeny; Retrotransposon.
© The Author(s). 2019.
Conflict of interest statement
Competing interestsThe authors declare that they have no competing interests.
Figures
Similar articles
-
A computational reconstruction of Papio phylogeny using Alu insertion polymorphisms.Mob DNA. 2018 Apr 5;9:13. doi: 10.1186/s13100-018-0118-3. eCollection 2018. Mob DNA. 2018. PMID: 29632618 Free PMC article.
-
Identification in gelada baboons (Theropithecus gelada) of a distinct simian T-cell lymphotropic virus type 3 with a broad range of Western blot reactivity.J Gen Virol. 2004 Feb;85(Pt 2):507-519. doi: 10.1099/vir.0.19630-0. J Gen Virol. 2004. PMID: 14769908
-
Fibrinopeptides A and B of baboons (Papio anubis, Papio hamadryas, and Theropithecus gelada): their amino acid sequences and evolutionary rates and a molecular phylogeny for the baboons.J Biochem. 1983 Dec;94(6):1973-8. doi: 10.1093/oxfordjournals.jbchem.a134551. J Biochem. 1983. PMID: 6423621
-
A proper study for mankind: Analogies from the Papionin monkeys and their implications for human evolution.Am J Phys Anthropol. 2001;Suppl 33:177-204. doi: 10.1002/ajpa.10021. Am J Phys Anthropol. 2001. PMID: 11786995 Review.
-
Theropithecus gelada distribution and variations related to taxonomy: history, challenges and implications for conservation.Primates. 2010 Oct;51(4):291-7. doi: 10.1007/s10329-010-0202-x. Epub 2010 May 28. Primates. 2010. PMID: 20509040 Review.
Cited by
-
Hybrid apes in the Anthropocene: Burden or asset for conservation?People Nat (Hoboken). 2021 Jun;3(3):573-586. doi: 10.1002/pan3.10214. Epub 2021 May 12. People Nat (Hoboken). 2021. PMID: 34805779 Free PMC article.
-
Genomic variation in baboons from central Mozambique unveils complex evolutionary relationships with other Papio species.BMC Ecol Evol. 2022 Apr 11;22(1):44. doi: 10.1186/s12862-022-01999-7. BMC Ecol Evol. 2022. PMID: 35410131 Free PMC article.
-
Analysis of Simian Endogenous Retrovirus (SERV) Full-Length Proviruses in Old World Monkey Genomes.Genes (Basel). 2022 Jan 10;13(1):119. doi: 10.3390/genes13010119. Genes (Basel). 2022. PMID: 35052460 Free PMC article.
-
A Comprehensive Overview of Baboon Phylogenetic History.Genes (Basel). 2023 Feb 28;14(3):614. doi: 10.3390/genes14030614. Genes (Basel). 2023. PMID: 36980887 Free PMC article. Review.
-
Insertion of Telomeric Repeats in the Human and Horse Genomes: An Evolutionary Perspective.Int J Mol Sci. 2020 Apr 18;21(8):2838. doi: 10.3390/ijms21082838. Int J Mol Sci. 2020. PMID: 32325780 Free PMC article.
References
-
- Geoffroy S-HI. Description des Mammifères Nouveaux ou Imparfaitement Connus de la Collection du Muséum d’Histoire Naturelle, et Remarques sur la Classification et les Caractères des Mammifères. Arch Mus Hist Nat Paris. 1843;2:485–592.
-
- Jablonski NG. Theropithecus: the rise and fall of a primate genus. Cambridge: Cambridge University press, 2005; 2005.
-
- Groves C. Primate taxonomy. Washington, DC: Smithsonian Institution Press; 2001.
-
- Jolly CJ. Species, subspecies and baboon systematics. In: LBM WHK, editor. Species, Species concepts and Primate Evolution. New York: Plenum Press; 1993. pp. 67–107.
Grants and funding
LinkOut - more resources
Full Text Sources

