Incomer, a DD36E family of Tc1/mariner transposons newly discovered in animals
- PMID: 31788035
- PMCID: PMC6875036
- DOI: 10.1186/s13100-019-0188-x
Incomer, a DD36E family of Tc1/mariner transposons newly discovered in animals
Abstract
Background: The Tc1/mariner superfamily might represent the most diverse and widely distributed group of DNA transposons. Several families have been identified; however, exploring the diversity of this superfamily and updating its classification is still ongoing in the life sciences.
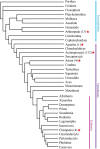
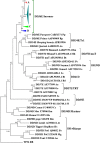
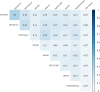
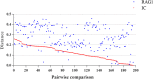
Results: Here we identified a new family of Tc1/mariner transposons, named Incomer (IC), which is close to, but distinct from the known family DD34E/Tc1. ICs have a total length of about 1.2 kb, and harbor a single open reading frame encoding a ~ 346 amino acid transposase with a DD36E motif and flanked by short terminal inverted repeats (TIRs) (22-32 base pairs, bp). This family is absent from prokaryotes, and is mainly distributed among vertebrates (141 species of four classes), including Agnatha (one species of jawless fish), Actinopterygii (132 species of ray-finned fish), Amphibia (four species of frogs), and Mammalia (four species of bats), but have a restricted distribution in invertebrates (four species in Insecta and nine in Arachnida). All ICs in bats (Myotis lucifugus, Eptesicus fuscus, Myotis davidii, and Myotis brandtii) are present as truncated copies in these genomes, and most of them are flanked by relatively long TIRs (51-126 bp). High copy numbers of miniature inverted-repeat transposable elements (MITEs) derived from ICs were also identified in bat genomes. Phylogenetic analysis revealed that ICs are more closely related to DD34E/Tc1 than to other families of Tc1/mariner (e.g., DD34D/mariner and DD × D/pogo), and can be classified into four distinct clusters. The host and IC phylogenies and pairwise distance comparisons between RAG1 genes and all consensus sequences of ICs support the idea that multiple episodes of horizontal transfer (HT) of ICs have occurred in vertebrates. In addition, the discovery of intact transposases, perfect TIRs and target site duplications of ICs suggests that this family may still be active in Insecta, Arachnida, frogs, and fish.
Conclusions: Exploring the diversity of Tc1/mariner transposons and revealing their evolutionary profiles will help provide a better understanding of the evolution of DNA transposons and their impact on genomic evolution. Here, a newly discovered family (DD36E/Incomer) of Tc1/mariner transposons is described in animals. It displays a similar structural organization and close relationship with the known DD34E/Tc1 elements, but has a relatively narrow distribution, indicating that DD36E/IC might have originated from the DD34E/Tc1 family. Our data also support the hypothesis of horizontal transfer of IC in vertebrates, even invading one lineage of mammals (bats). This study expands our understanding of the diversity of Tc1/mariner transposons and updates the classification of this superfamily.
Keywords: DD36E; Horizontal transfer; Tc1/mariner transposons.
© The Author(s). 2019.
Conflict of interest statement
Competing interestsThe authors declare that they have no competing interests.
Figures

Similar articles
-
Intruder (DD38E), a recently evolved sibling family of DD34E/Tc1 transposons in animals.Mob DNA. 2020 Dec 10;11(1):32. doi: 10.1186/s13100-020-00227-7. Mob DNA. 2020. PMID: 33303022 Free PMC article.
-
Divergent evolution profiles of DD37D and DD39D families of Tc1/mariner transposons in eukaryotes.Mol Phylogenet Evol. 2021 Aug;161:107143. doi: 10.1016/j.ympev.2021.107143. Epub 2021 Mar 10. Mol Phylogenet Evol. 2021. PMID: 33713798
-
Hiker, a new family of DNA transposons encoding transposases with DD35E motifs, displays a distinct phylogenetic relationship with most known DNA transposon families of IS630-Tc1-mariner (ITm).Mol Phylogenet Evol. 2023 Nov;188:107906. doi: 10.1016/j.ympev.2023.107906. Epub 2023 Aug 15. Mol Phylogenet Evol. 2023. PMID: 37586577
-
Research progress of Tc1/Mariner superfamily.Yi Chuan. 2017 Jan 20;39(1):1-13. doi: 10.16288/j.yczz.16-160. Yi Chuan. 2017. PMID: 28115300 Review.
-
Mariner and the ITm Superfamily of Transposons.Microbiol Spectr. 2015 Apr;3(2):MDNA3-0033-2014.. doi: 10.1128/microbiolspec.MDNA3-0033-2014. Microbiol Spectr. 2015. PMID: 26104691 Review.
Cited by
-
Multiple Invasions of Visitor, a DD41D Family of Tc1/mariner Transposons, throughout the Evolution of Vertebrates.Genome Biol Evol. 2020 Jul 1;12(7):1060-1073. doi: 10.1093/gbe/evaa135. Genome Biol Evol. 2020. PMID: 32602886 Free PMC article.
-
Intruder (DD38E), a recently evolved sibling family of DD34E/Tc1 transposons in animals.Mob DNA. 2020 Dec 10;11(1):32. doi: 10.1186/s13100-020-00227-7. Mob DNA. 2020. PMID: 33303022 Free PMC article.
-
Evolution of pogo, a separate superfamily of IS630-Tc1-mariner transposons, revealing recurrent domestication events in vertebrates.Mob DNA. 2020 Jul 22;11:25. doi: 10.1186/s13100-020-00220-0. eCollection 2020. Mob DNA. 2020. PMID: 32742312 Free PMC article.
-
Horizontal Transfer and Evolutionary Profiles of Two Tc1/DD34E Transposons (ZB and SB) in Vertebrates.Genes (Basel). 2022 Nov 29;13(12):2239. doi: 10.3390/genes13122239. Genes (Basel). 2022. PMID: 36553507 Free PMC article.
-
Prokaryotic and Eukaryotic Horizontal Transfer of Sailor (DD82E), a New Superfamily of IS630-Tc1-Mariner DNA Transposons.Biology (Basel). 2021 Oct 7;10(10):1005. doi: 10.3390/biology10101005. Biology (Basel). 2021. PMID: 34681104 Free PMC article.
References
LinkOut - more resources
Full Text Sources

