Highly sensitive MLH1 methylation analysis in blood identifies a cancer patient with low-level mosaic MLH1 epimutation
- PMID: 31779681
- PMCID: PMC6883525
- DOI: 10.1186/s13148-019-0762-6
Highly sensitive MLH1 methylation analysis in blood identifies a cancer patient with low-level mosaic MLH1 epimutation
Abstract
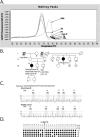
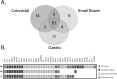
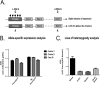
Constitutional MLH1 methylation (epimutation) is a rare cause of Lynch syndrome. Low-level methylation (≤ 10%) has occasionally been described. This study aimed to identify low-level constitutional MLH1 epimutations and determine its causal role in patients with MLH1-hypermethylated colorectal cancer.Eighteen patients with MLH1-hypermethylated colorectal tumors in whom MLH1 methylation was previously undetected in blood by methylation-specific multiplex ligation-dependent probe amplification (MS-MLPA) were screened for MLH1 methylation using highly sensitive MS-melting curve analysis (MS-MCA). Constitutional methylation was characterized by different approaches.MS-MCA identified one patient (5.6%) with low-level MLH1 methylation (~ 1%) in blood and other normal tissues, which was confirmed by clonal bisulfite sequencing in blood. The patient had developed three clonally related gastrointestinal MLH1-methylated tumor lesions at 22, 24, and 25 years of age. The methylated region in normal tissues overlapped with that reported for other carriers of constitutional MLH1 epimutations. Low-level MLH1 methylation and reduced allelic expression were linked to the same genetic haplotype, whereas the opposite allele was lost in patient's tumors. Mutation screening of MLH1 and other hereditary cancer genes was negative.Herein, a highly sensitive MS-MCA-based approach has demonstrated its utility for the identification of low-level constitutional MLH1 epigenetic mosaicism. The eventual identification and characterization of additional cases will be critical to ascertain the cancer risks associated with constitutional MLH1 epigenetic mosaicism.
Keywords: Constitutional MLH1 epimutation; Epigenetic mosaicism; Highly sensitive methodologies; Lynch syndrome; Methylation.
Conflict of interest statement
GC and JdV have received personal fees from VCN Biosciences and AstraZeneca, respectively, outside the submitted work. There are no other relationships or activities that could appear to have influenced the submitted work. All other authors declare that they have no competing interest.
Figures

Similar articles
-
MLH1 methylation screening is effective in identifying epimutation carriers.Eur J Hum Genet. 2012 Dec;20(12):1256-64. doi: 10.1038/ejhg.2012.136. Epub 2012 Jul 4. Eur J Hum Genet. 2012. PMID: 22763379 Free PMC article.
-
MLH1-methylated endometrial cancer under 60 years of age as the "sentinel" cancer in female carriers of high-risk constitutional MLH1 epimutation.Gynecol Oncol. 2023 Apr;171:129-140. doi: 10.1016/j.ygyno.2023.02.017. Epub 2023 Mar 8. Gynecol Oncol. 2023. PMID: 36893489 Free PMC article.
-
Finding the needle in a haystack: identification of cases of Lynch syndrome with MLH1 epimutation.Fam Cancer. 2016 Jul;15(3):413-22. doi: 10.1007/s10689-016-9887-3. Fam Cancer. 2016. PMID: 26886015 Review.
-
Contribution of MLH1 constitutional methylation for Lynch syndrome diagnosis in patients with tumor MLH1 downregulation.Cancer Med. 2018 Feb;7(2):433-444. doi: 10.1002/cam4.1285. Epub 2018 Jan 17. Cancer Med. 2018. PMID: 29341452 Free PMC article.
-
Lynch syndrome-associated endometrial carcinoma with MLH1 germline mutation and MLH1 promoter hypermethylation: a case report and literature review.BMC Cancer. 2018 May 21;18(1):576. doi: 10.1186/s12885-018-4489-0. BMC Cancer. 2018. PMID: 29783979 Free PMC article. Review.
Cited by
-
Assessment of tumor suppressor promoter methylation in healthy individuals.Clin Epigenetics. 2020 Aug 28;12(1):131. doi: 10.1186/s13148-020-00920-7. Clin Epigenetics. 2020. PMID: 32859265 Free PMC article.
-
Identifying primary and secondary MLH1 epimutation carriers displaying low-level constitutional MLH1 methylation using droplet digital PCR and genome-wide DNA methylation profiling of colorectal cancers.Clin Epigenetics. 2023 Jun 3;15(1):95. doi: 10.1186/s13148-023-01511-y. Clin Epigenetics. 2023. PMID: 37270516 Free PMC article.
-
Multifaceted Roles of DNA Methylation in Neoplastic Transformation, from Tumor Suppressors to EMT and Metastasis.Genes (Basel). 2020 Aug 12;11(8):922. doi: 10.3390/genes11080922. Genes (Basel). 2020. PMID: 32806509 Free PMC article. Review.
-
Unraveling noncoding DNA variants and epimutations: a paradigm shift in hereditary cancer research.Future Oncol. 2024;20(18):1289-1298. doi: 10.2217/fon-2023-0665. Epub 2024 May 9. Future Oncol. 2024. PMID: 38722139 Review.
-
Diagnosis of Lynch Syndrome and Strategies to Distinguish Lynch-Related Tumors from Sporadic MSI/dMMR Tumors.Cancers (Basel). 2021 Jan 26;13(3):467. doi: 10.3390/cancers13030467. Cancers (Basel). 2021. PMID: 33530449 Free PMC article. Review.

