Systematic prediction of FFAT motifs across eukaryote proteomes identifies nucleolar and eisosome proteins with the predicted capacity to form bridges to the endoplasmic reticulum
- PMID: 31777772
- PMCID: PMC6881177
- DOI: 10.1177/2515256419883136
Systematic prediction of FFAT motifs across eukaryote proteomes identifies nucleolar and eisosome proteins with the predicted capacity to form bridges to the endoplasmic reticulum
Abstract
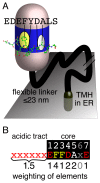
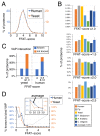
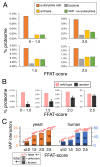
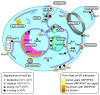
The endoplasmic reticulum (ER), the most pervasive organelle, exchanges information and material with many other organelles, but the extent of its inter-organelle connections and the proteins that form bridges are not well known. The integral ER membrane protein VAMP-associated protein (VAP) is found in multiple bridges, interacting with many proteins that contain a short linear motif consisting of "two phenylalanines in an acidic tract" (FFAT). The VAP-FFAT interaction is the most common mechanism by which cytoplasmic proteins, particularly inter-organelle bridges, target the ER. Therefore, predicting new FFAT motifs may both find new individual peripheral ER proteins and identify new routes of communication involving the ER. Here we searched for FFAT motifs across whole proteomes. The excess of eukaryotic proteins with FFAT motifs over background was ≥0.8%, suggesting this is the minimum number of peripheral ER proteins. In yeast, where VAP was previously known to bind 4 proteins with FFAT motifs, a detailed analysis of a subset of proteins predicted 20 FFAT motifs. Extrapolating these findings to the whole proteome estimated the number of FFAT motifs in yeast at approximately 50-55 (0.9% of proteome). Among these previously unstudied FFAT motifs, most have known functions outside the ER, so could be involved in inter-organelle communication. Many of these can target well-characterised membrane contact sites, however some are in nucleoli and eisosomes, organelles previously unknown to have molecular bridges to the ER. We speculate that the nucleolar and eisosomal proteins with predicted motifs may function while bridging to the ER, indicating novel ER-nucleolus and ER-eisosome routes of inter-organelle communication.
Keywords: BOP1; NOP2/Nsun6; UBR4; VPS13; eisosome; endoplasmic reticulum; lipid transfer protein; membrane contact sites; nucleolus; short linear motif.
Figures
Similar articles
-
VAP, a Versatile Access Point for the Endoplasmic Reticulum: Review and analysis of FFAT-like motifs in the VAPome.Biochim Biophys Acta. 2016 Aug;1861(8 Pt B):952-961. doi: 10.1016/j.bbalip.2016.02.009. Epub 2016 Feb 17. Biochim Biophys Acta. 2016. PMID: 26898182 Review.
-
Analysis of the key elements of FFAT-like motifs identifies new proteins that potentially bind VAP on the ER, including two AKAPs and FAPP2.PLoS One. 2012;7(1):e30455. doi: 10.1371/journal.pone.0030455. Epub 2012 Jan 19. PLoS One. 2012. PMID: 22276202 Free PMC article.
-
What the VAP: The Expanded VAP Family of Proteins Interacting With FFAT and FFAT-Related Motifs for Interorganellar Contact.Contact (Thousand Oaks). 2021 May 9;4:25152564211012246. doi: 10.1177/25152564211012246. eCollection 2021 Jan 1. Contact (Thousand Oaks). 2021. PMID: 34036242 Free PMC article.
-
FFAT motif phosphorylation controls formation and lipid transfer function of inter-organelle contacts.EMBO J. 2020 Dec 1;39(23):e104369. doi: 10.15252/embj.2019104369. Epub 2020 Oct 30. EMBO J. 2020. PMID: 33124732 Free PMC article.
-
The Interactome of the VAP Family of Proteins: An Overview.Cells. 2021 Jul 14;10(7):1780. doi: 10.3390/cells10071780. Cells. 2021. PMID: 34359948 Free PMC article. Review.
Cited by
-
In silico modeling human VPS13 proteins associated with donor and target membranes suggests lipid transfer mechanisms.Proteins. 2023 Apr;91(4):439-455. doi: 10.1002/prot.26446. Epub 2022 Dec 5. Proteins. 2023. PMID: 36404287 Free PMC article.
-
VPS13D bridges the ER to mitochondria and peroxisomes via Miro.J Cell Biol. 2021 May 3;220(5):e202010004. doi: 10.1083/jcb.202010004. J Cell Biol. 2021. PMID: 33891013 Free PMC article.
-
TRPC3 channel gating by lipids requires localization at the ER/PM junctions defined by STIM1.J Cell Biol. 2022 May 2;221(5):e202107120. doi: 10.1083/jcb.202107120. Epub 2022 Apr 13. J Cell Biol. 2022. PMID: 35416932 Free PMC article.
-
Interactions by Disorder - A Matter of Context.Front Mol Biosci. 2020 Jun 16;7:110. doi: 10.3389/fmolb.2020.00110. eCollection 2020. Front Mol Biosci. 2020. PMID: 32613009 Free PMC article. Review.
-
Systematic analysis of membrane contact sites in Saccharomyces cerevisiae uncovers modulators of cellular lipid distribution.Elife. 2022 Nov 10;11:e74602. doi: 10.7554/eLife.74602. Elife. 2022. PMID: 36354737 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
