Beyond reversal: ubiquitin and ubiquitin-like proteases and the orchestration of the DNA double strand break repair response
- PMID: 31769469
- PMCID: PMC6925521
- DOI: 10.1042/BST20190534
Beyond reversal: ubiquitin and ubiquitin-like proteases and the orchestration of the DNA double strand break repair response
Abstract
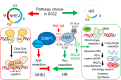
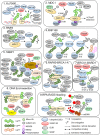
The cellular response to genotoxic DNA double strand breaks (DSBs) uses a multitude of post-translational modifications to localise, modulate and ultimately clear DNA repair factors in a timely and accurate manner. Ubiquitination is well established as vital to the DSB response, with a carefully co-ordinated pathway of histone ubiquitination events being a central component of DSB signalling. Other ubiquitin-like modifiers (Ubl) including SUMO and NEDD8 have since been identified as playing important roles in DSB repair. In the last five years ∼20 additional Ub/Ubl proteases have been implicated in the DSB response. The number of proteases identified highlights the complexity of the Ub/Ubl signal present at DSBs. Ub/Ubl proteases regulate turnover, activity and protein-protein interactions of DSB repair factors both catalytically and non-catalytically. This not only ensures efficient repair of breaks but has a role in channelling repair into the correct DSB repair sub-pathways. Ultimately Ub/Ubl proteases have essential roles in maintaining genomic stability. Given that deficiencies in many Ub/Ubl proteases promotes sensitivity to DNA damaging chemotherapies, they could be attractive targets for cancer treatment.
Keywords: DNA synthesis and repair; DUB; SENP; double strand break; sumoylation; ubiquitin.
© 2019 The Author(s).
Conflict of interest statement
The Author declares that there are no competing interests associated with this manuscript.
Figures
Similar articles
-
SUMO in the DNA Double-Stranded Break Response: Similarities, Differences, and Cooperation with Ubiquitin.J Mol Biol. 2017 Nov 10;429(22):3376-3387. doi: 10.1016/j.jmb.2017.05.012. Epub 2017 May 17. J Mol Biol. 2017. PMID: 28527786 Review.
-
Regulation of DNA double-strand break repair by ubiquitin and ubiquitin-like modifiers.Nat Rev Mol Cell Biol. 2016 May 23;17(6):379-94. doi: 10.1038/nrm.2016.58. Nat Rev Mol Cell Biol. 2016. PMID: 27211488 Review.
-
The Lys63-deubiquitylating Enzyme BRCC36 Limits DNA Break Processing and Repair.J Biol Chem. 2016 Jul 29;291(31):16197-207. doi: 10.1074/jbc.M116.731927. Epub 2016 Jun 10. J Biol Chem. 2016. PMID: 27288411 Free PMC article.
-
RNF4, a SUMO-targeted ubiquitin E3 ligase, promotes DNA double-strand break repair.Genes Dev. 2012 Jun 1;26(11):1179-95. doi: 10.1101/gad.188284.112. Genes Dev. 2012. PMID: 22661229 Free PMC article.
-
Ubiquitylation, neddylation and the DNA damage response.Open Biol. 2015 Apr;5(4):150018. doi: 10.1098/rsob.150018. Open Biol. 2015. PMID: 25833379 Free PMC article. Review.
Cited by
-
Erasing marks: Functions of plant deubiquitylating enzymes in modulating the ubiquitin code.Plant Cell. 2024 Sep 3;36(9):3057-3073. doi: 10.1093/plcell/koae129. Plant Cell. 2024. PMID: 38656977 Review.
-
E3 ligases: a ubiquitous link between DNA repair, DNA replication and human disease.Biochem J. 2024 Jul 17;481(14):923-944. doi: 10.1042/BCJ20240124. Biochem J. 2024. PMID: 38985307 Free PMC article. Review.
-
Advancements in colorectal cancer research: Unveiling the cellular and molecular mechanisms of neddylation (Review).Int J Oncol. 2024 Apr;64(4):39. doi: 10.3892/ijo.2024.5627. Epub 2024 Feb 23. Int J Oncol. 2024. PMID: 38391033 Free PMC article. Review.
-
Arginine methylation and ubiquitylation crosstalk controls DNA end-resection and homologous recombination repair.Nat Commun. 2021 Nov 2;12(1):6313. doi: 10.1038/s41467-021-26413-6. Nat Commun. 2021. PMID: 34728620 Free PMC article.
-
Contribution of proteases to the hallmarks of aging and to age-related neurodegeneration.Aging Cell. 2022 May;21(5):e13603. doi: 10.1111/acel.13603. Epub 2022 Mar 29. Aging Cell. 2022. PMID: 35349763 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

