The ENCODE Portal as an Epigenomics Resource
- PMID: 31751002
- PMCID: PMC7307447
- DOI: 10.1002/cpbi.89
The ENCODE Portal as an Epigenomics Resource
Abstract
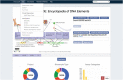
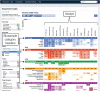
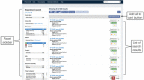
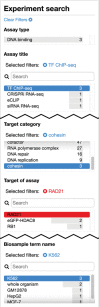
The Encyclopedia of DNA Elements (ENCODE) web portal hosts genomic data generated by the ENCODE Consortium, Genomics of Gene Regulation, The NIH Roadmap Epigenomics Consortium, and the modENCODE and modERN projects. The goal of the ENCODE project is to build a comprehensive map of the functional elements of the human and mouse genomes. Currently, the portal database stores over 500 TB of raw and processed data from over 15,000 experiments spanning assays that measure gene expression, DNA accessibility, DNA and RNA binding, DNA methylation, and 3D chromatin structure across numerous cell lines, tissue types, and differentiation states with selected genetic and molecular perturbations. The ENCODE portal provides unrestricted access to the aforementioned data and relevant metadata as a service to the scientific community. The metadata model captures the details of the experiments, raw and processed data files, and processing pipelines in human and machine-readable form and enables the user to search for specific data either using a web browser or programmatically via REST API. Furthermore, ENCODE data can be freely visualized or downloaded for additional analyses. © 2019 The Authors. Basic Protocol: Query the portal Support Protocol 1: Batch downloading Support Protocol 2: Using the cart to download files Support Protocol 3: Visualize data Alternate Protocol: Query building and programmatic access.
Keywords: ENCODE; database; epigenetics; human genome; regulatory elements.
© 2019 The Authors.
Figures

Similar articles
-
The Encyclopedia of DNA elements (ENCODE): data portal update.Nucleic Acids Res. 2018 Jan 4;46(D1):D794-D801. doi: 10.1093/nar/gkx1081. Nucleic Acids Res. 2018. PMID: 29126249 Free PMC article.
-
New developments on the Encyclopedia of DNA Elements (ENCODE) data portal.Nucleic Acids Res. 2020 Jan 8;48(D1):D882-D889. doi: 10.1093/nar/gkz1062. Nucleic Acids Res. 2020. PMID: 31713622 Free PMC article.
-
ENCODE data at the ENCODE portal.Nucleic Acids Res. 2016 Jan 4;44(D1):D726-32. doi: 10.1093/nar/gkv1160. Epub 2015 Nov 2. Nucleic Acids Res. 2016. PMID: 26527727 Free PMC article.
-
Ontology application and use at the ENCODE DCC.Database (Oxford). 2015 Mar 16;2015:bav010. doi: 10.1093/database/bav010. Print 2015. Database (Oxford). 2015. PMID: 25776021 Free PMC article. Review.
-
Using the ENCODE Resource for Functional Annotation of Genetic Variants.Cold Spring Harb Protoc. 2015 Mar 11;2015(6):522-36. doi: 10.1101/pdb.top084988. Cold Spring Harb Protoc. 2015. PMID: 25762420 Free PMC article. Review.
Cited by
-
Transcriptome-wide association analysis identifies DACH1 as a kidney disease risk gene that contributes to fibrosis.J Clin Invest. 2021 May 17;131(10):e141801. doi: 10.1172/JCI141801. J Clin Invest. 2021. PMID: 33998598 Free PMC article.
-
Bromodomain Inhibition Attenuates the Progression and Sensitizes the Chemosensitivity of Osteosarcoma by Repressing GP130/STAT3 Signaling.Front Oncol. 2021 Jun 8;11:642134. doi: 10.3389/fonc.2021.642134. eCollection 2021. Front Oncol. 2021. PMID: 34168981 Free PMC article.
-
echolocatoR: an automated end-to-end statistical and functional genomic fine-mapping pipeline.Bioinformatics. 2022 Jan 3;38(2):536-539. doi: 10.1093/bioinformatics/btab658. Bioinformatics. 2022. PMID: 34529038 Free PMC article.
-
Representing core gene expression activity relationships using the latent structure implicit in Bayesian networks.Bioinformatics. 2024 Aug 2;40(8):btae463. doi: 10.1093/bioinformatics/btae463. Bioinformatics. 2024. PMID: 39051682 Free PMC article.
-
Heterochromatin rewiring and domain disruption-mediated chromatin compaction during erythropoiesis.Nat Struct Mol Biol. 2023 Apr;30(4):463-474. doi: 10.1038/s41594-023-00939-3. Epub 2023 Mar 13. Nat Struct Mol Biol. 2023. PMID: 36914797
References
Literature Cited
Internet Resources
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

