Structural and Biochemical Investigations of the [4Fe-4S] Cluster-Containing Fumarate Hydratase from Leishmania major
- PMID: 31743022
- PMCID: PMC7065722
- DOI: 10.1021/acs.biochem.9b00923
Structural and Biochemical Investigations of the [4Fe-4S] Cluster-Containing Fumarate Hydratase from Leishmania major
Abstract
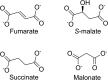
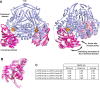
Class I fumarate hydratases (FHs) are central metabolic enzymes that use a [4Fe-4S] cluster to catalyze the reversible conversion of fumarate to S-malate. The parasite Leishmania major, which is responsible for leishmaniasis, expresses two class I FH isoforms: mitochondrial LmFH-1 and cytosolic LmFH-2. In this study, we present kinetic characterizations of both LmFH isoforms, present 13 crystal structures of LmFH-2 variants, and employ site-directed mutagenesis to investigate the enzyme's mechanism. Our kinetic data confirm that both LmFH-1 and LmFH-2 are susceptible to oxygen-dependent inhibition, with data from crystallography and electron paramagnetic resonance spectroscopy showing that oxygen exposure converts an active [4Fe-4S] cluster to an inactive [3Fe-4S] cluster. Our anaerobically conducted kinetic studies reveal a preference for fumarate over S-malate. Our data further reveal that single alanine substitutions of T467, R421, R471, D135, and H334 decrease kcat values 9-16000-fold without substantially affecting Km values, suggesting that these residues function in catalytic roles. Crystal structures of LmFH-2 variants are consistent with this idea, showing similar bidentate binding to the unique iron of the [4Fe-4S] cluster for substrate S-malate as observed in wild type FH. We further present LmFH-2 structures with substrate fumarate and weak inhibitors succinate and malonate bound in the active site and the first structure of an LmFH that is substrate-free and inhibitor-free, the latter showing increased mobility in the C-terminal domain. Collectively, these data provide insight into the molecular basis for the reaction catalyzed by LmFHs, enzymes that are potential drug targets against leishmaniasis.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
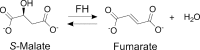



Similar articles
-
Crystal structure of an Fe-S cluster-containing fumarate hydratase enzyme from Leishmania major reveals a unique protein fold.Proc Natl Acad Sci U S A. 2016 Aug 30;113(35):9804-9. doi: 10.1073/pnas.1605031113. Epub 2016 Aug 15. Proc Natl Acad Sci U S A. 2016. PMID: 27528683 Free PMC article.
-
Fumarate hydratase isoforms of Leishmania major: subcellular localization, structural and kinetic properties.Int J Biol Macromol. 2012 Jul-Aug;51(1-2):25-31. doi: 10.1016/j.ijbiomac.2012.04.025. Epub 2012 May 5. Int J Biol Macromol. 2012. PMID: 22569531
-
Crystal Structures of Fumarate Hydratases from Leishmania major in a Complex with Inhibitor 2-Thiomalate.ACS Chem Biol. 2019 Feb 15;14(2):266-275. doi: 10.1021/acschembio.8b00972. Epub 2019 Jan 24. ACS Chem Biol. 2019. PMID: 30645090 Free PMC article.
-
Oxygen sensing by the global regulator, FNR: the role of the iron-sulfur cluster.FEMS Microbiol Rev. 1998 Dec;22(5):341-52. doi: 10.1111/j.1574-6976.1998.tb00375.x. FEMS Microbiol Rev. 1998. PMID: 9990723 Review.
-
Reactions of nitric oxide and oxygen with the regulator of fumarate and nitrate reduction, a global transcriptional regulator, during anaerobic growth of Escherichia coli.Methods Enzymol. 2008;437:191-209. doi: 10.1016/S0076-6879(07)37011-0. Methods Enzymol. 2008. PMID: 18433630 Review.
Cited by
-
Cyanobacterial Dihydroxyacid Dehydratases Are a Promising Growth Inhibition Target.ACS Chem Biol. 2020 Aug 21;15(8):2281-2288. doi: 10.1021/acschembio.0c00507. Epub 2020 Aug 12. ACS Chem Biol. 2020. PMID: 32786290 Free PMC article.
-
The thiolation of uridine 34 in tRNA, which controls protein translation, depends on a [4Fe-4S] cluster in the archaeum Methanococcus maripaludis.Sci Rep. 2023 Apr 1;13(1):5351. doi: 10.1038/s41598-023-32423-9. Sci Rep. 2023. PMID: 37005440 Free PMC article.
References
-
- Coustou V.; Besteiro S.; Riviere L.; Biran M.; Biteau N.; Franconi J. M.; Boshart M.; Baltz T.; Bringaud F. (2005) A mitochondrial NADH-dependent fumarate reductase involved in the production of succinate excreted by procyclic Trypanosoma brucei. J. Biol. Chem. 280, 16559–16570. 10.1074/jbc.M500343200. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

