Next-generation sequencing of dsRNA is greatly improved by treatment with the inexpensive denaturing reagent DMSO
- PMID: 31738702
- PMCID: PMC6927307
- DOI: 10.1099/mgen.0.000315
Next-generation sequencing of dsRNA is greatly improved by treatment with the inexpensive denaturing reagent DMSO
Abstract
dsRNA is the genetic material of important viruses and a key component of RNA interference-based immunity in eukaryotes. Previous studies have noted difficulties in determining the sequence of dsRNA molecules that have affected studies of immune function and estimates of viral diversity in nature. DMSO has been used to denature dsRNA prior to the reverse-transcription stage to improve reverse transcriptase PCR and Sanger sequencing. We systematically tested the utility of DMSO to improve the sequencing yield of a dsRNA virus (Φ6) in a short-read next-generation sequencing platform. DMSO treatment improved sequencing read recovery by over two orders of magnitude, even when RNA and cDNA concentrations were below the limit of detection. We also tested the effects of DMSO on a mock eukaryotic viral community and found that dsRNA virus reads increased with DMSO treatment. Furthermore, we provide evidence that DMSO treatment does not adversely affect recovery of reads from a ssRNA viral genome (influenza A/California/07/2009). We suggest that up to 50 % DMSO treatment be used prior to cDNA synthesis when samples of interest are composed of or may contain dsRNA.
Keywords: dsRNA; dsRNAseq; innate immunity; metagenomics; sociovirology; viral sequencing.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures

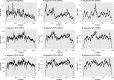
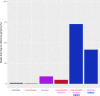

Similar articles
-
RNA Viral Metagenome Analysis of Subnanogram dsRNA Using Fragmented and Primer Ligated dsRNA Sequencing (FLDS).Microbes Environ. 2021;36(2):ME20152. doi: 10.1264/jsme2.ME20152. Microbes Environ. 2021. PMID: 33952860 Free PMC article.
-
The large genome segment of dsRNA bacteriophage phi6 is the key regulator in the in vitro minus and plus strand synthesis.RNA. 1995 Jul;1(5):510-8. RNA. 1995. PMID: 7489512 Free PMC article.
-
FLDS: A Comprehensive dsRNA Sequencing Method for Intracellular RNA Virus Surveillance.Microbes Environ. 2016;31(1):33-40. doi: 10.1264/jsme2.ME15171. Epub 2016 Feb 13. Microbes Environ. 2016. PMID: 26877136 Free PMC article.
-
Reverse genetics of dsRNA bacteriophage phi 6.Adv Virus Res. 1999;53:341-53. doi: 10.1016/s0065-3527(08)60355-3. Adv Virus Res. 1999. PMID: 10582106 Review. No abstract available.
-
Utilization of Bacteriophage phi6 for the Production of High-Quality Double-Stranded RNA Molecules.Viruses. 2024 Jan 22;16(1):166. doi: 10.3390/v16010166. Viruses. 2024. PMID: 38275976 Free PMC article. Review.
Cited by
-
Unveiling Viruses Associated with Gastroenteritis Using a Metagenomics Approach.Viruses. 2020 Dec 13;12(12):1432. doi: 10.3390/v12121432. Viruses. 2020. PMID: 33322135 Free PMC article.
-
Comparison of Three Viral Nucleic Acid Preamplification Pipelines for Sewage Viral Metagenomics.Food Environ Virol. 2024 Sep;16(3):1-22. doi: 10.1007/s12560-024-09594-3. Epub 2024 Apr 22. Food Environ Virol. 2024. PMID: 38647859 Free PMC article.
-
Identification and characterization of porcine Rotavirus A in Chilean swine population.Front Vet Sci. 2023 Nov 2;10:1240346. doi: 10.3389/fvets.2023.1240346. eCollection 2023. Front Vet Sci. 2023. PMID: 38026647 Free PMC article.
-
Three modes of viral adaption by the heart.Sci Adv. 2024 Nov 15;10(46):eadp6303. doi: 10.1126/sciadv.adp6303. Epub 2024 Nov 13. Sci Adv. 2024. PMID: 39536108 Free PMC article.
-
RNA Viruses in Aquatic Ecosystems through the Lens of Ecological Genomics and Transcriptomics.Viruses. 2022 Mar 28;14(4):702. doi: 10.3390/v14040702. Viruses. 2022. PMID: 35458432 Free PMC article. Review.

