Human NORs, comprising rDNA arrays and functionally conserved distal elements, are located within dynamic chromosomal regions
- PMID: 31727772
- PMCID: PMC6942050
- DOI: 10.1101/gad.331892.119
Human NORs, comprising rDNA arrays and functionally conserved distal elements, are located within dynamic chromosomal regions
Abstract
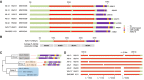
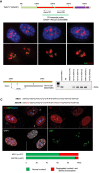
Human nucleolar organizer regions (NORs), containing ribosomal gene (rDNA) arrays, are located on the p-arms of acrocentric chromosomes (HSA13-15, 21, and 22). Absence of these p-arms from genome references has hampered research on nucleolar formation. Previously, we assembled a distal junction (DJ) DNA sequence contig that abuts rDNA arrays on their telomeric side, revealing that it is shared among the acrocentrics and impacts nucleolar organization. To facilitate inclusion into genome references, we describe sequencing the DJ from all acrocentrics, including three versions of HSA21, ∼3 Mb of novel sequence. This was achieved by exploiting monochromosomal somatic cell hybrids containing single human acrocentric chromosomes with NORs that retain functional potential. Analyses revealed remarkable DJ sequence and functional conservation among human acrocentrics. Exploring chimpanzee acrocentrics, we show that "DJ-like" sequences and abutting rDNA arrays are inverted as a unit in comparison to humans. Thus, rDNA arrays and linked DJs represent a conserved functional locus. We provide direct evidence for exchanges between heterologous human acrocentric p-arms, and uncover extensive structural variation between chromosomes and among individuals. These findings lead us to revaluate the molecular definition of NORs, identify novel genomic structural variation, and provide a rationale for the distinctive chromosomal organization of NORs.
Keywords: acrocentric chromosomes; nucleolar organizer regions (NORs); nucleolus; recombination; ribosomal DNA.
© 2019 van Sluis; Published by Cold Spring Harbor Laboratory Press.
Figures


Comment in
-
The human nucleolus organizer regions.Genes Dev. 2019 Dec 1;33(23-24):1617-1618. doi: 10.1101/gad.334748.119. Genes Dev. 2019. PMID: 31792016 Free PMC article. Review.
Similar articles
-
NORs on human acrocentric chromosome p-arms are active by default and can associate with nucleoli independently of rDNA.Proc Natl Acad Sci U S A. 2020 May 12;117(19):10368-10377. doi: 10.1073/pnas.2001812117. Epub 2020 Apr 24. Proc Natl Acad Sci U S A. 2020. PMID: 32332163 Free PMC article.
-
Actively transcribed rDNA and distal junction (DJ) sequence are involved in association of NORs with nucleoli.Cell Mol Life Sci. 2023 Apr 12;80(5):121. doi: 10.1007/s00018-023-04770-3. Cell Mol Life Sci. 2023. PMID: 37043028 Free PMC article.
-
Nucleolar organizer regions: genomic 'dark matter' requiring illumination.Genes Dev. 2016 Jul 15;30(14):1598-610. doi: 10.1101/gad.283838.116. Genes Dev. 2016. PMID: 27474438 Free PMC article. Review.
-
The p-Arms of Human Acrocentric Chromosomes Play by a Different Set of Rules.Annu Rev Genomics Hum Genet. 2023 Aug 25;24:63-83. doi: 10.1146/annurev-genom-101122-081642. Epub 2023 Feb 28. Annu Rev Genomics Hum Genet. 2023. PMID: 36854315 Review.
-
Nucleolar organization, ribosomal DNA array stability, and acrocentric chromosome integrity are linked to telomere function.PLoS One. 2014 Mar 24;9(3):e92432. doi: 10.1371/journal.pone.0092432. eCollection 2014. PLoS One. 2014. PMID: 24662969 Free PMC article.
Cited by
-
Recombination between heterologous human acrocentric chromosomes.Nature. 2023 May;617(7960):335-343. doi: 10.1038/s41586-023-05976-y. Epub 2023 May 10. Nature. 2023. PMID: 37165241 Free PMC article.
-
Topological stress triggers persistent DNA lesions in ribosomal DNA with ensuing formation of PML-nucleolar compartment.Elife. 2024 Oct 10;12:RP91304. doi: 10.7554/eLife.91304. Elife. 2024. PMID: 39388244 Free PMC article.
-
MicroRNAs and long non-coding RNAs as novel regulators of ribosome biogenesis.Biochem Soc Trans. 2020 Apr 29;48(2):595-612. doi: 10.1042/BST20190854. Biochem Soc Trans. 2020. PMID: 32267487 Free PMC article. Review.
-
Crossing boundaries of light microscopy resolution discerns novel assemblies in the nucleolus.Histochem Cell Biol. 2024 Jul;162(1-2):161-183. doi: 10.1007/s00418-024-02297-7. Epub 2024 May 17. Histochem Cell Biol. 2024. PMID: 38758429 Free PMC article.
-
Monoallelically expressed noncoding RNAs form nucleolar territories on NOR-containing chromosomes and regulate rRNA expression.Elife. 2024 Jan 19;13:e80684. doi: 10.7554/eLife.80684. Elife. 2024. PMID: 38240312 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
