Global Vectors Representation of Protein Sequences and Its Application for Predicting Self-Interacting Proteins with Multi-Grained Cascade Forest Model
- PMID: 31726752
- PMCID: PMC6896115
- DOI: 10.3390/genes10110924
Global Vectors Representation of Protein Sequences and Its Application for Predicting Self-Interacting Proteins with Multi-Grained Cascade Forest Model
Abstract
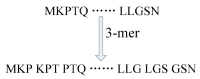
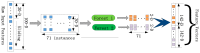
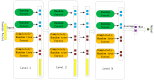
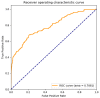
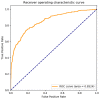
Self-interacting proteins (SIPs) is of paramount importance in current molecular biology. There have been developed a number of traditional biological experiment methods for predicting SIPs in the past few years. However, these methods are costly, time-consuming and inefficient, and often limit their usage for predicting SIPs. Therefore, the development of computational method emerges at the times require. In this paper, we for the first time proposed a novel deep learning model which combined natural language processing (NLP) method for potential SIPs prediction from the protein sequence information. More specifically, the protein sequence is de novo assembled by k-mers. Then, we obtained the global vectors representation for each protein sequences by using natural language processing (NLP) technique. Finally, based on the knowledge of known self-interacting and non-interacting proteins, a multi-grained cascade forest model is trained to predict SIPs. Comprehensive experiments were performed on yeast and human datasets, which obtained an accuracy rate of 91.45% and 93.12%, respectively. From our evaluations, the experimental results show that the use of amino acid semantics information is very helpful for addressing the problem of sequences containing both self-interacting and non-interacting pairs of proteins. This work would have potential applications for various biological classification problems.
Keywords: de novo protein sequence; global vector representation; multi-grained cascade forest; self-interacting proteins.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Predicting protein-protein interactions from primary protein sequences using a novel multi-scale local feature representation scheme and the random forest.PLoS One. 2015 May 6;10(5):e0125811. doi: 10.1371/journal.pone.0125811. eCollection 2015. PLoS One. 2015. PMID: 25946106 Free PMC article.
-
PSPEL: In Silico Prediction of Self-Interacting Proteins from Amino Acids Sequences Using Ensemble Learning.IEEE/ACM Trans Comput Biol Bioinform. 2017 Sep-Oct;14(5):1165-1172. doi: 10.1109/TCBB.2017.2649529. Epub 2017 Jan 10. IEEE/ACM Trans Comput Biol Bioinform. 2017. PMID: 28092572
-
Robust and accurate prediction of self-interacting proteins from protein sequence information by exploiting weighted sparse representation based classifier.BMC Bioinformatics. 2022 Dec 1;23(Suppl 7):518. doi: 10.1186/s12859-022-04880-y. BMC Bioinformatics. 2022. PMID: 36457083 Free PMC article.
-
Computational Models for Self-Interacting Proteins Prediction.Protein Pept Lett. 2020;27(5):392-399. doi: 10.2174/0929866527666191227141713. Protein Pept Lett. 2020. PMID: 31880240 Review.
-
A survey on computational models for predicting protein-protein interactions.Brief Bioinform. 2021 Sep 2;22(5):bbab036. doi: 10.1093/bib/bbab036. Brief Bioinform. 2021. PMID: 33693513 Review.
Cited by
-
MFIDMA: A Multiple Information Integration Model for the Prediction of Drug-miRNA Associations.Biology (Basel). 2022 Dec 26;12(1):41. doi: 10.3390/biology12010041. Biology (Basel). 2022. PMID: 36671734 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous