Dissect the DNMT3A- and DNMT3B-mediated DNA Co-methylation through a Covalent Complex Approach
- PMID: 31726062
- PMCID: PMC6995754
- DOI: 10.1016/j.jmb.2019.11.004
Dissect the DNMT3A- and DNMT3B-mediated DNA Co-methylation through a Covalent Complex Approach
Abstract
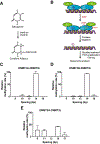
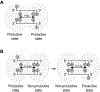
DNA methylation plays a critical role in regulating gene expression, genomic stability, and cell fate commitment. Mammalian DNA methylation, which mostly occurs in the context of CpG dinucleotide, is installed by two denovo DNA methyltransferases, DNMT3A and DNMT3B. Oligomerization of DNMT3A and DNMT3B permits both enzymes to comethylate two CpG sites located on the same DNA substrates. However, how DNMT3A- and DNMT3B-mediated co-methylation contributes to the DNA methylation patterns remain unclear. Here we generated covalent enzyme-substrate complexes of DNMT3A and DNMT3B, and performed bisulfite sequencing-based single-turnover methylation analysis on both complexes. Our results showed that both DNMT3A- and DNMT3B-mediated co-methylation preferentially gives rise to a methylation spacing of 14 base pairs, consistent with the previous structural observation for DNMT3A in complex with regulatory protein DNMT3L and CpG DNA. This study provides a novel method for mechanistic investigation of DNMT3A- and DNMT3B-mediated DNA co-methylation.
Keywords: Covalent complex; CpG spacing; DNA methylation; DNMT3A; DNMT3B.
Copyright © 2019 Elsevier Ltd. All rights reserved.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

Similar articles
-
CpG sites preferentially methylated by Dnmt3a in vivo.J Biol Chem. 2006 Apr 14;281(15):9901-8. doi: 10.1074/jbc.M511100200. Epub 2006 Jan 26. J Biol Chem. 2006. PMID: 16439359
-
Structural basis for DNMT3A-mediated de novo DNA methylation.Nature. 2018 Feb 15;554(7692):387-391. doi: 10.1038/nature25477. Epub 2018 Feb 7. Nature. 2018. PMID: 29414941 Free PMC article.
-
DNMT3L stimulates the DNA methylation activity of Dnmt3a and Dnmt3b through a direct interaction.J Biol Chem. 2004 Jun 25;279(26):27816-23. doi: 10.1074/jbc.M400181200. Epub 2004 Apr 21. J Biol Chem. 2004. PMID: 15105426
-
Genetic Studies on Mammalian DNA Methyltransferases.Adv Exp Med Biol. 2016;945:123-150. doi: 10.1007/978-3-319-43624-1_6. Adv Exp Med Biol. 2016. PMID: 27826837 Review.
-
Molecular enzymology of mammalian DNA methyltransferases.Curr Top Microbiol Immunol. 2006;301:203-25. doi: 10.1007/3-540-31390-7_7. Curr Top Microbiol Immunol. 2006. PMID: 16570849 Review.
Cited by
-
Specific DNMT3C flanking sequence preferences facilitate methylation of young murine retrotransposons.Commun Biol. 2024 May 16;7(1):582. doi: 10.1038/s42003-024-06252-z. Commun Biol. 2024. PMID: 38755427 Free PMC article.
-
MicroRNA-29a inhibits cell proliferation and arrests cell cycle by modulating p16 methylation in cervical cancer.Oncol Lett. 2021 Apr;21(4):272. doi: 10.3892/ol.2021.12533. Epub 2021 Feb 9. Oncol Lett. 2021. PMID: 33717269 Free PMC article.
-
Long Non-coding RNA IRAIN Inhibits VEGFA Expression via Enhancing Its DNA Methylation Leading to Tumor Suppression in Renal Carcinoma.Front Oncol. 2020 Sep 2;10:1082. doi: 10.3389/fonc.2020.01082. eCollection 2020. Front Oncol. 2020. PMID: 32983957 Free PMC article.
-
Enzymology of Mammalian DNA Methyltransferases.Adv Exp Med Biol. 2022;1389:69-110. doi: 10.1007/978-3-031-11454-0_4. Adv Exp Med Biol. 2022. PMID: 36350507
-
Beta class amino methyltransferases from bacteria to humans: evolution and structural consequences.Nucleic Acids Res. 2020 Oct 9;48(18):10034-10044. doi: 10.1093/nar/gkaa446. Nucleic Acids Res. 2020. PMID: 32453412 Free PMC article. Review.
References
-
- Bourc’his D & Bestor TH Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 431, 96–99 (2004). - PubMed
-
- Holliday R & Pugh JE DNA modification mechanisms and gene activity during development. Science 187, 226–232 (1975). - PubMed
-
- Walsh CP, Chaillet JR & Bestor TH Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat Genet 20, 116–117 (1998). - PubMed
-
- Li E, Beard C & Jaenisch R Role for DNA methylation in genomic imprinting. Nature 366, 362–365 (1993). - PubMed
-
- Panning B & Jaenisch R RNA and the epigenetic regulation of X chromosome inactivation. Cell 93, 305–308 (1998). - PubMed

