Impact of Antiretroviral Therapy Duration on HIV-1 Infection of T Cells within Anatomic Sites
- PMID: 31723024
- PMCID: PMC7000983
- DOI: 10.1128/JVI.01270-19
Impact of Antiretroviral Therapy Duration on HIV-1 Infection of T Cells within Anatomic Sites
Abstract
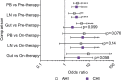
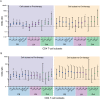
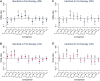
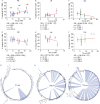
Understanding the impact of antiretroviral therapy (ART) duration on HIV-infected cells is critical for developing successful curative strategies. To address this issue, we conducted a cross-sectional/inter-participant genetic characterization of HIV-1 RNA from pre- and on-therapy plasmas and HIV-1 DNA from CD4+ T cell subsets derived from peripheral blood (PB), lymph node (LN), and gut tissues of 26 participants after 3 to 17.8 years of ART. Our studies revealed in four acute/early participants who had paired PB and LN samples a substantial reduction in the proportion of HIV-infected cells per year on therapy within the LN. Extrapolation to all 12 acute/early participants estimated a much smaller reduction in the proportion of HIV-1-infected cells within LNs per year on therapy that was similar to that in the participants treated during chronic infection. LN-derived effector memory T (TEM) cells contained HIV-1 DNA that was genetically identical to viral sequences derived from pre- and on-therapy plasma samples. The proportion of identical HIV-1 DNA sequences increased within PB-derived TEM cells. However, the infection frequency of TEM cells in PB was stable, indicating that cellular proliferation that compensates for T cell loss over time contributes to HIV-1 persistence. This study suggests that ART reduces HIV-infected T cells and that clonal expansion of HIV-infected cells maintains viral persistence. Importantly, LN-derived TEM cells are a probable source of HIV-1 genomes capable of producing infectious HIV-1 and should be targeted by future curative strategies.IMPORTANCE HIV-1 persists as an integrated genome in CD4+ memory T cells during effective therapy, and cessation of current treatments results in resumption of viral replication. To date, the impact of antiretroviral therapy duration on HIV-infected CD4+ T cells and the mechanisms of viral persistence in different anatomic sites is not clearly elucidated. In the current study, we found that treatment duration was associated with a reduction in HIV-infected T cells. Our genetic analyses revealed that CD4+ effector memory T (TEM) cells derived from the lymph node appeared to contain provirus that was genetically identical to plasma-derived virions. Moreover, we found that cellular proliferation counterbalanced the decay of HIV-infected cells throughout therapy. The contribution of cellular proliferation to viral persistence is particularly significant in TEM cells. Our study emphasizes the importance of HIV-1 intervention and provides new insights into the location of memory T cells infected with HIV-1 DNA, which is capable of contributing to viremia.
Keywords: CD4+ T cell subsets; HIV-1; HIV-1 persistence; acute/early infection; anatomic sites; cellular proliferation; chronic infection; long-term antiretroviral therapy; single-genome sequencing; single-proviral sequencing.
Copyright © 2020 Lee et al.
Figures







Similar articles
-
Genetic Diversity, Compartmentalization, and Age of HIV Proviruses Persisting in CD4+ T Cell Subsets during Long-Term Combination Antiretroviral Therapy.J Virol. 2020 Feb 14;94(5):e01786-19. doi: 10.1128/JVI.01786-19. Print 2020 Feb 14. J Virol. 2020. PMID: 31776273 Free PMC article.
-
Lack of concordance between residual viremia and viral variants driving de novo infection of CD4(+) T cells on ART.Retrovirology. 2016 Aug 2;13(1):51. doi: 10.1186/s12977-016-0282-9. Retrovirology. 2016. PMID: 27484989 Free PMC article.
-
Peripheral T Follicular Helper Cells Are the Major HIV Reservoir within Central Memory CD4 T Cells in Peripheral Blood from Chronically HIV-Infected Individuals on Combination Antiretroviral Therapy.J Virol. 2015 Dec 16;90(6):2718-28. doi: 10.1128/JVI.02883-15. J Virol. 2015. PMID: 26676775 Free PMC article. Clinical Trial.
-
Single-molecule techniques to quantify and genetically characterise persistent HIV.Retrovirology. 2018 Jan 9;15(1):3. doi: 10.1186/s12977-017-0386-x. Retrovirology. 2018. PMID: 29316955 Free PMC article. Review.
-
The role of integration and clonal expansion in HIV infection: live long and prosper.Retrovirology. 2018 Oct 23;15(1):71. doi: 10.1186/s12977-018-0448-8. Retrovirology. 2018. PMID: 30352600 Free PMC article. Review.
Cited by
-
Plasma-Derived HIV-1 Virions Contain Considerable Levels of Defective Genomes.J Virol. 2022 Mar 23;96(6):e0201121. doi: 10.1128/jvi.02011-21. Epub 2022 Mar 23. J Virol. 2022. PMID: 35201897 Free PMC article.
-
Unequal distribution of genetically-intact HIV-1 proviruses in cells expressing the immune checkpoint markers PD-1 and/or CTLA-4.Front Immunol. 2023 Jan 26;14:1064346. doi: 10.3389/fimmu.2023.1064346. eCollection 2023. Front Immunol. 2023. PMID: 36776833 Free PMC article.
-
Morphine exposure exacerbates HIV-1 Tat driven changes to neuroinflammatory factors in cultured astrocytes.PLoS One. 2020 Mar 25;15(3):e0230563. doi: 10.1371/journal.pone.0230563. eCollection 2020. PLoS One. 2020. PMID: 32210470 Free PMC article.
-
Plasmacytoid dendritic cells have divergent effects on HIV infection of initial target cells and induce a pro-retention phenotype.PLoS Pathog. 2021 Apr 19;17(4):e1009522. doi: 10.1371/journal.ppat.1009522. eCollection 2021 Apr. PLoS Pathog. 2021. PMID: 33872331 Free PMC article.
-
Assessment of HIV-1 integration in tissues and subsets across infection stages.JCI Insight. 2020 Oct 15;5(20):e139783. doi: 10.1172/jci.insight.139783. JCI Insight. 2020. PMID: 32970634 Free PMC article.
References
-
- Chun TW, Carruth L, Finzi D, Shen X, DiGiuseppe JA, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn TC, Kuo YH, Brookmeyer R, Zeiger MA, Barditch-Crovo P, Siliciano RF. 1997. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 387:183–188. doi:10.1038/387183a0. - DOI - PubMed
-
- Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, Quinn TC, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho DD, Richman DD, Siliciano RF. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295–1300. doi:10.1126/science.278.5341.1295. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

