Puumala and Andes Orthohantaviruses Cause Transient Protein Kinase R-Dependent Formation of Stress Granules
- PMID: 31723021
- PMCID: PMC7000972
- DOI: 10.1128/JVI.01168-19
Puumala and Andes Orthohantaviruses Cause Transient Protein Kinase R-Dependent Formation of Stress Granules
Abstract
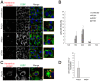
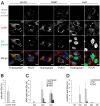
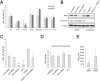
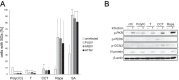
Virus infection frequently triggers host cell stress signaling resulting in translational arrest; as a consequence, many viruses employ means to modulate the host stress response. Hantaviruses are negative-sense, single-stranded RNA viruses known to inhibit host innate immune responses and apoptosis, but their impact on host cell stress signaling remains largely unknown. In this study, we investigated activation of host cell stress responses during hantavirus infection. We show that hantavirus infection causes transient formation of stress granules (SGs) but does so in only a limited proportion of infected cells. Our data indicate some cell type-specific and hantavirus species-specific variability in SG prevalence and show SG formation to be dependent on the activation of protein kinase R (PKR). Hantavirus infection inhibited PKR-dependent SG formation, which could account for the transient nature and low prevalence of SG formation observed during hantavirus infection. In addition, we report only limited colocalization of hantaviral proteins or RNA with SGs and show evidence indicating hantavirus-mediated inhibition of PKR-like endoplasmic reticulum (ER) kinase (PERK).IMPORTANCE Our work presents the first report on stress granule formation during hantavirus infection. We show that hantavirus infection actively inhibits stress granule formation, thereby escaping the detrimental effects on global translation imposed by host stress signaling. Our results highlight a previously uncharacterized aspect of hantavirus-host interactions with possible implications for how hantaviruses are able to cause persistent infection in natural hosts and for pathogenesis.
Keywords: PKR; hantavirus; stress granule.
Copyright © 2020 American Society for Microbiology.
Figures

Similar articles
-
Andes virus nucleocapsid protein interrupts protein kinase R dimerization to counteract host interference in viral protein synthesis.J Virol. 2015 Feb;89(3):1628-39. doi: 10.1128/JVI.02347-14. Epub 2014 Nov 19. J Virol. 2015. PMID: 25410857 Free PMC article.
-
The PERK/PKR-eIF2α Pathway Negatively Regulates Porcine Hemagglutinating Encephalomyelitis Virus Replication by Attenuating Global Protein Translation and Facilitating Stress Granule Formation.J Virol. 2022 Jan 12;96(1):e0169521. doi: 10.1128/JVI.01695-21. Epub 2021 Oct 13. J Virol. 2022. PMID: 34643429 Free PMC article.
-
Mouse Norovirus Infection Arrests Host Cell Translation Uncoupled from the Stress Granule-PKR-eIF2α Axis.mBio. 2019 Jun 18;10(3):e00960-19. doi: 10.1128/mBio.00960-19. mBio. 2019. PMID: 31213553 Free PMC article.
-
What Do We Know about How Hantaviruses Interact with Their Different Hosts?Viruses. 2016 Aug 11;8(8):223. doi: 10.3390/v8080223. Viruses. 2016. PMID: 27529272 Free PMC article. Review.
-
Hantavirus-induced immunity in rodent reservoirs and humans.Immunol Rev. 2008 Oct;225:163-89. doi: 10.1111/j.1600-065X.2008.00694.x. Immunol Rev. 2008. PMID: 18837782 Review.
Cited by
-
Diverse susceptibilities and responses of human and rodent cells to orthohantavirus infection reveal different levels of cellular restriction.PLoS Negl Trop Dis. 2022 Oct 12;16(10):e0010844. doi: 10.1371/journal.pntd.0010844. eCollection 2022 Oct. PLoS Negl Trop Dis. 2022. PMID: 36223391 Free PMC article.
-
Stress Granules as Causes and Consequences of Translation Suppression.Antioxid Redox Signal. 2023 Aug;39(4-6):390-409. doi: 10.1089/ars.2022.0164. Epub 2023 Jun 28. Antioxid Redox Signal. 2023. PMID: 37183403 Free PMC article. Review.
-
Innate immune response against vector-borne bunyavirus infection and viral countermeasures.Front Cell Infect Microbiol. 2024 Apr 22;14:1365221. doi: 10.3389/fcimb.2024.1365221. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 38711929 Free PMC article. Review.
-
Viruses Run: The Evasion Mechanisms of the Antiviral Innate Immunity by Hantavirus.Front Microbiol. 2021 Sep 30;12:759198. doi: 10.3389/fmicb.2021.759198. eCollection 2021. Front Microbiol. 2021. PMID: 34659193 Free PMC article. Review.
-
Innate Immunity to Orthohantaviruses: Could Divergent Immune Interactions Explain Host-specific Disease Outcomes?J Mol Biol. 2022 Mar 30;434(6):167230. doi: 10.1016/j.jmb.2021.167230. Epub 2021 Sep 4. J Mol Biol. 2022. PMID: 34487792 Free PMC article. Review.
References
-
- Klingström J, Smed-Sörensen A, Maleki KT, Solà-Riera C, Ahlm C, Björkström NK, Ljunggren HG. 2019. Innate and adaptive immune responses against human Puumala virus infection: immunopathogenesis and suggestions for novel treatment strategies for severe hantavirus-associated syndromes. J Intern Med 285:510–523. doi:10.1111/joim.12876. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

