Transcriptomic Signatures Predict Regulators of Drug Synergy and Clinical Regimen Efficacy against Tuberculosis
- PMID: 31719182
- PMCID: PMC6851285
- DOI: 10.1128/mBio.02627-19
Transcriptomic Signatures Predict Regulators of Drug Synergy and Clinical Regimen Efficacy against Tuberculosis
Abstract
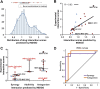
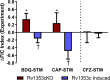
The rapid spread of multidrug-resistant strains has created a pressing need for new drug regimens to treat tuberculosis (TB), which kills 1.8 million people each year. Identifying new regimens has been challenging due to the slow growth of the pathogen Mycobacterium tuberculosis (MTB), coupled with the large number of possible drug combinations. Here we present a computational model (INDIGO-MTB) that identified synergistic regimens featuring existing and emerging anti-TB drugs after screening in silico more than 1 million potential drug combinations using MTB drug transcriptomic profiles. INDIGO-MTB further predicted the gene Rv1353c as a key transcriptional regulator of multiple drug interactions, and we confirmed experimentally that Rv1353c upregulation reduces the antagonism of the bedaquiline-streptomycin combination. A retrospective analysis of 57 clinical trials of TB regimens using INDIGO-MTB revealed that synergistic combinations were significantly more efficacious than antagonistic combinations (P value = 1 × 10-4) based on the percentage of patients with negative sputum cultures after 8 weeks of treatment. Our study establishes a framework for rapid assessment of TB drug combinations and is also applicable to other bacterial pathogens.IMPORTANCE Multidrug combination therapy is an important strategy for treating tuberculosis, the world's deadliest bacterial infection. Long treatment durations and growing rates of drug resistance have created an urgent need for new approaches to prioritize effective drug regimens. Hence, we developed a computational model called INDIGO-MTB that identifies synergistic drug regimens from an immense set of possible drug combinations using the pathogen response transcriptome elicited by individual drugs. Although the underlying input data for INDIGO-MTB was generated under in vitro broth culture conditions, the predictions from INDIGO-MTB correlated significantly with in vivo drug regimen efficacy from clinical trials. INDIGO-MTB also identified the transcription factor Rv1353c as a regulator of multiple drug interaction outcomes, which could be targeted for rationally enhancing drug synergy.
Keywords: Mycobacterium tuberculosis; computer modeling; drug combinations; drug interactions; drug synergy; gene expression; transcription factors; transcriptomics; tuberculosis.
Copyright © 2019 Ma et al.
Figures

Similar articles
-
Harnessing Biological Insight to Accelerate Tuberculosis Drug Discovery.Acc Chem Res. 2019 Aug 20;52(8):2340-2348. doi: 10.1021/acs.accounts.9b00275. Epub 2019 Jul 30. Acc Chem Res. 2019. PMID: 31361123 Free PMC article. Review.
-
Comparison of Two Modern Survival Prediction Tools, SORG-MLA and METSSS, in Patients With Symptomatic Long-bone Metastases Who Underwent Local Treatment With Surgery Followed by Radiotherapy and With Radiotherapy Alone.Clin Orthop Relat Res. 2024 Dec 1;482(12):2193-2208. doi: 10.1097/CORR.0000000000003185. Epub 2024 Jul 23. Clin Orthop Relat Res. 2024. PMID: 39051924
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Ceftazidime with avibactam for treating severe aerobic Gram-negative bacterial infections: technology evaluation to inform a novel subscription-style payment model.Health Technol Assess. 2024 Oct;28(73):1-230. doi: 10.3310/YAPL9347. Health Technol Assess. 2024. PMID: 39487661 Free PMC article.
-
Falls prevention interventions for community-dwelling older adults: systematic review and meta-analysis of benefits, harms, and patient values and preferences.Syst Rev. 2024 Nov 26;13(1):289. doi: 10.1186/s13643-024-02681-3. Syst Rev. 2024. PMID: 39593159 Free PMC article.
Cited by
-
The pursuit of mechanism of action: uncovering drug complexity in TB drug discovery.RSC Chem Biol. 2021 Apr 1;2(2):423-440. doi: 10.1039/d0cb00226g. Epub 2021 Jan 13. RSC Chem Biol. 2021. PMID: 33928253 Free PMC article.
-
The past, present and future of tuberculosis treatment.Zhejiang Da Xue Xue Bao Yi Xue Ban. 2022 Dec 25;51(6):657-668. doi: 10.3724/zdxbyxb-2022-0454. Zhejiang Da Xue Xue Bao Yi Xue Ban. 2022. PMID: 36915970 Free PMC article. Review. English.
-
Both Pharmacokinetic Variability and Granuloma Heterogeneity Impact the Ability of the First-Line Antibiotics to Sterilize Tuberculosis Granulomas.Front Pharmacol. 2020 Mar 24;11:333. doi: 10.3389/fphar.2020.00333. eCollection 2020. Front Pharmacol. 2020. PMID: 32265707 Free PMC article.
-
Wollamide Cyclic Hexapeptides Synergize with Established and New Tuberculosis Antibiotics in Targeting Mycobacterium tuberculosis.Microbiol Spectr. 2023 Aug 17;11(4):e0046523. doi: 10.1128/spectrum.00465-23. Epub 2023 Jun 8. Microbiol Spectr. 2023. PMID: 37289062 Free PMC article.
-
Pre-Clinical Tools for Predicting Drug Efficacy in Treatment of Tuberculosis.Microorganisms. 2022 Feb 26;10(3):514. doi: 10.3390/microorganisms10030514. Microorganisms. 2022. PMID: 35336089 Free PMC article. Review.
References
-
- World Health Organization. 2016. Global tuberculosis report 2016. World Health Organization, Geneva, Switzerland.
-
- Zumla A, Chakaya J, Centis R, D’Ambrosio L, Mwaba P, Bates M, Kapata N, Nyirenda T, Chanda D, Mfinanga S, Hoelscher M, Maeurer M, Migliori GB. 2015. Tuberculosis treatment and management–an update on treatment regimens, trials, new drugs, and adjunct therapies. Lancet Respir Med 3:220–234. doi:10.1016/S2213-2600(15)00063-6. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

