Circ-HuR suppresses HuR expression and gastric cancer progression by inhibiting CNBP transactivation
- PMID: 31718709
- PMCID: PMC6852727
- DOI: 10.1186/s12943-019-1094-z
Circ-HuR suppresses HuR expression and gastric cancer progression by inhibiting CNBP transactivation
Erratum in
-
Correction: Circ-HuR suppresses HuR expression and gastric cancer progression by inhibiting CNBP transactivation.Mol Cancer. 2023 Sep 20;22(1):155. doi: 10.1186/s12943-023-01862-3. Mol Cancer. 2023. PMID: 37730600 Free PMC article. No abstract available.
Abstract
Background: Circular RNAs (circRNAs), a subclass of non-coding RNAs, play essential roles in tumorigenesis and aggressiveness. Our previous study has identified that circAGO2 drives gastric cancer progression through activating human antigen R (HuR), a protein stabilizing AU-rich element-containing mRNAs. However, the functions and underlying mechanisms of circRNAs derived from HuR in gastric cancer progression remain elusive.
Methods: CircRNAs derived from HuR were detected by real-time quantitative RT-PCR and validated by Sanger sequencing. Biotin-labeled RNA pull-down, mass spectrometry, RNA immunoprecipitation, RNA electrophoretic mobility shift, and in vitro binding assays were applied to identify proteins interacting with circRNA. Gene expression regulation was observed by chromatin immunoprecipitation, dual-luciferase assay, real-time quantitative RT-PCR, and western blot assays. Gain- and loss-of-function studies were performed to observe the impacts of circRNA and its protein partner on the growth, invasion, and metastasis of gastric cancer cells in vitro and in vivo.
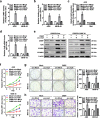
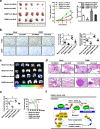
Results: Circ-HuR (hsa_circ_0049027) was predominantly detected in the nucleus, and was down-regulated in gastric cancer tissues and cell lines. Ectopic expression of circ-HuR suppressed the growth, invasion, and metastasis of gastric cancer cells in vitro and in vivo. Mechanistically, circ-HuR interacted with CCHC-type zinc finger nucleic acid binding protein (CNBP), and subsequently restrained its binding to HuR promoter, resulting in down-regulation of HuR and repression of tumor progression.
Conclusions: Circ-HuR serves as a tumor suppressor to inhibit CNBP-facilitated HuR expression and gastric cancer progression, indicating a potential therapeutic target for gastric cancer.
Keywords: CCHC-type zinc finger nucleic acid binding protein; Circular RNAs; Gastric cancer; Human antigen R.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




Similar articles
-
circ-hnRNPU inhibits NONO-mediated c-Myc transactivation and mRNA stabilization essential for glycosylation and cancer progression.J Exp Clin Cancer Res. 2023 Nov 23;42(1):313. doi: 10.1186/s13046-023-02898-5. J Exp Clin Cancer Res. 2023. PMID: 37993881 Free PMC article.
-
CircGSK3B promotes RORA expression and suppresses gastric cancer progression through the prevention of EZH2 trans-inhibition.J Exp Clin Cancer Res. 2021 Oct 19;40(1):330. doi: 10.1186/s13046-021-02136-w. J Exp Clin Cancer Res. 2021. PMID: 34666800 Free PMC article.
-
Exosomal circSHKBP1 promotes gastric cancer progression via regulating the miR-582-3p/HUR/VEGF axis and suppressing HSP90 degradation.Mol Cancer. 2020 Jun 29;19(1):112. doi: 10.1186/s12943-020-01208-3. Mol Cancer. 2020. PMID: 32600329 Free PMC article.
-
Drug delivery approaches for HuR-targeted therapy for lung cancer.Adv Drug Deliv Rev. 2022 Jan;180:114068. doi: 10.1016/j.addr.2021.114068. Epub 2021 Nov 22. Adv Drug Deliv Rev. 2022. PMID: 34822926 Free PMC article. Review.
-
Human antigen R: Exploring its inflammatory response impact and significance in cardiometabolic disorders.J Cell Physiol. 2024 May;239(5):e31229. doi: 10.1002/jcp.31229. Epub 2024 Mar 1. J Cell Physiol. 2024. PMID: 38426269 Review.
Cited by
-
Back to the Origin: Mechanisms of circRNA-Directed Regulation of Host Genes in Human Disease.Noncoding RNA. 2024 Sep 24;10(5):49. doi: 10.3390/ncrna10050049. Noncoding RNA. 2024. PMID: 39452835 Free PMC article. Review.
-
Circular RNAs as diagnostic biomarkers for gastric cancer: A comprehensive update from emerging functions to clinical significances.Front Genet. 2022 Oct 28;13:1037120. doi: 10.3389/fgene.2022.1037120. eCollection 2022. Front Genet. 2022. PMID: 36386850 Free PMC article. Review.
-
CircMAST1 inhibits cervical cancer progression by hindering the N4-acetylcytidine modification of YAP mRNA.Cell Mol Biol Lett. 2024 Feb 8;29(1):25. doi: 10.1186/s11658-024-00540-6. Cell Mol Biol Lett. 2024. PMID: 38331765 Free PMC article.
-
CircPVT1 promotes proliferation of lung squamous cell carcinoma by binding to miR-30d/e.J Exp Clin Cancer Res. 2021 Jun 10;40(1):193. doi: 10.1186/s13046-021-01976-w. J Exp Clin Cancer Res. 2021. PMID: 34112238 Free PMC article.
-
CircNEIL3 regulatory loop promotes pancreatic ductal adenocarcinoma progression via miRNA sponging and A-to-I RNA-editing.Mol Cancer. 2021 Mar 9;20(1):51. doi: 10.1186/s12943-021-01333-7. Mol Cancer. 2021. PMID: 33750389 Free PMC article.
References
-
- Doller A, el-S A, Huwiler A, Muller R, Radeke HH, Pfeilschifter J, Eberhardt W. Posttranslational modification of the AU-rich element binding protein HuR by protein kinase Cdelta elicits angiotensin II-induced stabilization and nuclear export of cyclooxygenase 2 mRNA. Mol Cell Biol. 2008;28:2608–2625. doi: 10.1128/MCB.01530-07. - DOI - PMC - PubMed
-
- Vazquez-Chantada M, Fernandez-Ramos D, Embade N, Martinez-Lopez N, Varela-Rey M, Woodhoo A, Luka Z, Wagner C, Anglim PP, Finnell RH, et al. HuR/methyl-HuR and AUF1 regulate the MAT expressed during liver proliferation, differentiation, and carcinogenesis. Gastroenterology. 2010;138:1943–1953. doi: 10.1053/j.gastro.2010.01.032. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

