Insights into the Effects of Cancer Associated Mutations at the UPF2 and ATP-Binding Sites of NMD Master Regulator: UPF1
- PMID: 31718065
- PMCID: PMC6888360
- DOI: 10.3390/ijms20225644
Insights into the Effects of Cancer Associated Mutations at the UPF2 and ATP-Binding Sites of NMD Master Regulator: UPF1
Abstract
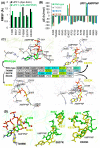
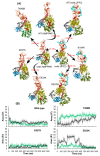
Nonsense-mediated mRNA decay (NMD) is a quality control mechanism that recognizes post-transcriptionally abnormal transcripts and mediates their degradation. The master regulator of NMD is UPF1, an enzyme with intrinsic ATPase and helicase activities. The cancer genomic sequencing data has identified frequently mutated residues in the CH-domain and ATP-binding site of UPF1. In silico screening of UPF1 stability change as a function over 41 cancer mutations has identified five variants with significant effects: K164R, R253W, T499M, E637K, and E833K. To explore the effects of these mutations on the associated energy landscape of UPF1, molecular dynamics simulations (MDS) were performed. MDS identified stable H-bonds between residues S152, S203, S205, Q230/R703, and UPF2/AMPPNP, and suggest that phosphorylation of Serine residues may control UPF1-UPF2 binding. Moreover, the alleles K164R and R253W in the CH-domain improved UPF1-UPF2 binding. In addition, E637K and E833K alleles exhibited improved UPF1-AMPPNP binding compared to the T499M variant; the lower binding is predicted from hindrance caused by the side-chain of T499M to the docking of the tri-phosphate moiety (AMPPNP) into the substrate site. The dynamics of wild-type/mutant systems highlights the flexible nature of the ATP-binding region in UPF1. These insights can facilitate the development of drug discovery strategies for manipulating NMD signaling in cell systems using chemical tools.
Keywords: ATP-binding site; UPF1; UPF2; cancer mutations; molecular dynamics simulations; structural stability.
Conflict of interest statement
The authors declare no competing interests.
Figures






Similar articles
-
NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity.Nat Struct Mol Biol. 2008 Jan;15(1):85-93. doi: 10.1038/nsmb1330. Epub 2007 Dec 9. Nat Struct Mol Biol. 2008. PMID: 18066079
-
Tight intramolecular regulation of the human Upf1 helicase by its N- and C-terminal domains.Nucleic Acids Res. 2013 Feb 1;41(4):2404-15. doi: 10.1093/nar/gks1320. Epub 2012 Dec 28. Nucleic Acids Res. 2013. PMID: 23275559 Free PMC article.
-
UPF1 helicase orchestrates mutually exclusive interactions with the SMG6 endonuclease and UPF2.Nucleic Acids Res. 2024 Jun 10;52(10):6036-6048. doi: 10.1093/nar/gkae323. Nucleic Acids Res. 2024. PMID: 38709891 Free PMC article.
-
UPFront and center in RNA decay: UPF1 in nonsense-mediated mRNA decay and beyond.RNA. 2019 Apr;25(4):407-422. doi: 10.1261/rna.070136.118. Epub 2019 Jan 17. RNA. 2019. PMID: 30655309 Free PMC article. Review.
-
Upf proteins: highly conserved factors involved in nonsense mRNA mediated decay.Mol Biol Rep. 2018 Feb;45(1):39-55. doi: 10.1007/s11033-017-4139-7. Epub 2017 Dec 27. Mol Biol Rep. 2018. PMID: 29282598 Review.
Cited by
-
Sensing of H2O2-induced oxidative stress by the UPF factor complex is crucial for activation of catalase-3 expression in Neurospora.PLoS Genet. 2023 Oct 16;19(10):e1010985. doi: 10.1371/journal.pgen.1010985. eCollection 2023 Oct. PLoS Genet. 2023. PMID: 37844074 Free PMC article.
-
The biological behavior of tRNA-derived fragment tRF-Leu-AAG in pancreatic cancer cells.Bioengineered. 2022 Apr;13(4):10617-10628. doi: 10.1080/21655979.2022.2064206. Bioengineered. 2022. PMID: 35442152 Free PMC article.
-
In Vitro Cross-Linking MS Reveals SMG1-UPF2-SMG7 Assembly as Molecular Partners within the NMD Surveillance.Int J Mol Sci. 2024 Mar 10;25(6):3182. doi: 10.3390/ijms25063182. Int J Mol Sci. 2024. PMID: 38542156 Free PMC article.
-
DNA and RNA Binding Proteins: From Motifs to Roles in Cancer.Int J Mol Sci. 2022 Aug 18;23(16):9329. doi: 10.3390/ijms23169329. Int J Mol Sci. 2022. PMID: 36012592 Free PMC article. Review.
-
Highly Conserved Homotrimer Cavity Formed by the SARS-CoV-2 Spike Glycoprotein: A Novel Binding Site.J Clin Med. 2020 May 14;9(5):1473. doi: 10.3390/jcm9051473. J Clin Med. 2020. PMID: 32422996 Free PMC article.
References
-
- Kashima I., Yamashita A., Izumi N., Kataoka N., Morishita R., Hoshino S., Ohno M., Dreyfuss G., Ohno S. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 2006;20:355–367. doi: 10.1101/gad.1389006. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

