Conditional Reprogramming for Patient-Derived Cancer Models and Next-Generation Living Biobanks
- PMID: 31717887
- PMCID: PMC6912808
- DOI: 10.3390/cells8111327
Conditional Reprogramming for Patient-Derived Cancer Models and Next-Generation Living Biobanks
Abstract
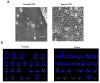
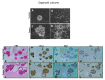
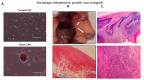
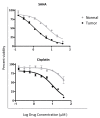
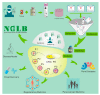
Traditional cancer models including cell lines and animal models have limited applications in both basic and clinical cancer research. Genomics-based precision oncology only help 2-20% patients with solid cancer. Functional diagnostics and patient-derived cancer models are needed for precision cancer biology. In this review, we will summarize applications of conditional cell reprogramming (CR) in cancer research and next generation living biobanks (NGLB). Together with organoids, CR has been cited in two NCI (National Cancer Institute, USA) programs (PDMR: patient-derived cancer model repository; HCMI: human cancer model initiatives. HCMI will be distributed through ATCC). Briefly, the CR method is a simple co-culture technology with a Rho kinase inhibitor, Y-27632, in combination with fibroblast feeder cells, which allows us to rapidly expand both normal and malignant epithelial cells from diverse anatomic sites and mammalian species and does not require transfection with exogenous viral or cellular genes. Establishment of CR cells from both normal and tumor tissue is highly efficient. The robust nature of the technique is exemplified by the ability to produce 2 × 106 cells in five days from a core biopsy of tumor tissue. Normal CR cell cultures retain a normal karyotype and differentiation potential and CR cells derived from tumors retain their tumorigenic phenotype. CR also allows us to enrich cancer cells from urine (for bladder cancer), blood (for prostate cancer), and pleural effusion (for non-small cell lung carcinoma). The ability to produce inexhaustible cell populations using CR technology from small biopsies and cryopreserved specimens has the potential to transform biobanking repositories (NGLB: next-generation living biobank) and current pathology practice by enabling genetic, biochemical, metabolomic, proteomic, and biological assays, including chemosensitivity testing as a functional diagnostics tool for precision cancer medicine. We discussed analyses of patient-derived matched normal and tumor models using a case with tongue squamous cell carcinoma as an example. Last, we summarized applications in cancer research, disease modeling, drug discovery, and regenerative medicine of CR-based NGLB.
Keywords: conditionally reprogrammed cells; living biobanks; organoids; patient-derived cancer models.
Conflict of interest statement
Several patents for conditional reprogramming technology has been awarded to Georgetown University by the United States Patent Office. The license for this technology has been given to Propagenix for commercialization. The inventors, R.S., X.L., and Georgetown University receive potential royalties and payments from Propagenix.
Figures
Similar articles
-
ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells.Am J Pathol. 2012 Feb;180(2):599-607. doi: 10.1016/j.ajpath.2011.10.036. Epub 2011 Dec 18. Am J Pathol. 2012. PMID: 22189618 Free PMC article.
-
Conditionally reprogrammed normal and primary tumor prostate epithelial cells: a novel patient-derived cell model for studies of human prostate cancer.Oncotarget. 2017 Apr 4;8(14):22741-22758. doi: 10.18632/oncotarget.13937. Oncotarget. 2017. PMID: 28009986 Free PMC article.
-
Conditional reprogramming: next generation cell culture.Acta Pharm Sin B. 2020 Aug;10(8):1360-1381. doi: 10.1016/j.apsb.2020.01.011. Epub 2020 Jan 26. Acta Pharm Sin B. 2020. PMID: 32963937 Free PMC article. Review.
-
Conditional cell reprogramming for modeling host-virus interactions and human viral diseases.J Med Virol. 2020 Nov;92(11):2440-2452. doi: 10.1002/jmv.26093. Epub 2020 Jun 16. J Med Virol. 2020. PMID: 32478897 Free PMC article. Review.
-
Conditionally reprogrammed cells (CRC) methodology does not allow the in vitro expansion of patient-derived primary and metastatic lung cancer cells.Int J Cancer. 2018 Jul 1;143(1):88-99. doi: 10.1002/ijc.31260. Epub 2018 Jan 26. Int J Cancer. 2018. PMID: 29341112
Cited by
-
Rapid screening for individualized chemotherapy optimization of colorectal cancer: A novel conditional reprogramming technology-based functional diagnostic assay.Transl Oncol. 2021 Jan;14(1):100935. doi: 10.1016/j.tranon.2020.100935. Epub 2020 Nov 12. Transl Oncol. 2021. PMID: 33190042 Free PMC article.
-
Personalized drug testing in a patient with non-small-cell lung cancer using cultured cancer cells from pleural effusion.J Int Med Res. 2020 Sep;48(9):300060520955058. doi: 10.1177/0300060520955058. J Int Med Res. 2020. PMID: 32954884 Free PMC article.
-
Insights into the characteristics of primary radioresistant cervical cancer using single-cell transcriptomics.Hum Cell. 2023 May;36(3):1135-1146. doi: 10.1007/s13577-023-00882-x. Epub 2023 Mar 3. Hum Cell. 2023. PMID: 36867313 Free PMC article.
-
Application of Organoid Models in Prostate Cancer Research.Front Oncol. 2021 Sep 27;11:736431. doi: 10.3389/fonc.2021.736431. eCollection 2021. Front Oncol. 2021. PMID: 34646778 Free PMC article. Review.
-
Establishment, characterization, and drug screening of low-passage patient individual non-small cell lung cancer in vitro models including the rare pleomorphic subentity.Front Oncol. 2023 May 9;13:1089681. doi: 10.3389/fonc.2023.1089681. eCollection 2023. Front Oncol. 2023. PMID: 37228492 Free PMC article.
References
-
- Daniel V.C., Marchionni L., Hierman J.S., Rhodes J.T., Devereux W.L., Rudin C.M., Yung R., Parmigiani G., Dorsch M., Peacock C.D., et al. A primary xenograft model of small-cell lung cancer reveals irreversible changes in gene expression imposed by culture in vitro. Cancer Res. 2009;69:3364–3373. doi: 10.1158/0008-5472.CAN-08-4210. - DOI - PMC - PubMed
-
- McDermott U., Sharma S.V., Dowell L., Greninger P., Montagut C., Lamb J., Archibald H., Raudales R., Tam A., Lee D., et al. Identification of genotype-correlated sensitivity to selective kinase inhibitors by using high-throughput tumor cell line profiling. Proc. Natl. Acad. Sci. USA. 2007;104:19936–19941. doi: 10.1073/pnas.0707498104. - DOI - PMC - PubMed
-
- Barretina J., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehar J., Kryukov G.V., Sonkin D., et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. - DOI - PMC - PubMed
-
- Johnson J.I., Decker S., Zaharevitz D., Rubinstein L.V., Venditti J.M., Schepartz S., Kalyandrug S., Christian M., Arbuck S., Hollingshead M., et al. Relationships between drug activity in NCI preclinical in vitro and in vivo models and early clinical trials. Br. J. Cancer. 2001;84:1424–1431. doi: 10.1054/bjoc.2001.1796. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

