IVIVC Assessment of Two Mouse Brain Endothelial Cell Models for Drug Screening
- PMID: 31717321
- PMCID: PMC6920823
- DOI: 10.3390/pharmaceutics11110587
IVIVC Assessment of Two Mouse Brain Endothelial Cell Models for Drug Screening
Erratum in
-
Erratum: Puscas, I.; et al. IVIVC Assessment of Two Mouse Brain Endothelial Cell Models for Drug Screening. Pharmaceutics 2019, 11, 587.Pharmaceutics. 2020 Jun 4;12(6):514. doi: 10.3390/pharmaceutics12060514. Pharmaceutics. 2020. PMID: 32512770 Free PMC article.
Abstract
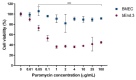
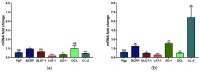
Since most preclinical drug permeability assays across the blood-brain barrier (BBB) are still evaluated in rodents, we compared an in vitro mouse primary endothelial cell model to the mouse b.End3 and the acellular parallel artificial membrane permeability assay (PAMPA) models for drug screening purposes. The mRNA expression of key feature membrane proteins of primary and bEnd.3 mouse brain endothelial cells were compared. Transwell® monolayer models were further characterized in terms of tightness and integrity. The in vitro in vivo correlation (IVIVC) was obtained by the correlation of the in vitro permeability data with log BB values obtained in mice for seven drugs. The mouse primary model showed higher monolayer integrity and levels of mRNA expression of BBB tight junction (TJ) proteins and membrane transporters (MBRT), especially for the efflux transporter Pgp. The IVIVC and drug ranking underlined the superiority of the primary model (r2 = 0.765) when compared to the PAMPA-BBB (r2 = 0.391) and bEnd.3 cell line (r2 = 0.019) models. The primary monolayer mouse model came out as a simple and reliable candidate for the prediction of drug permeability across the BBB. This model encompasses a rapid set-up, a fair reproduction of BBB tissue characteristics, and an accurate drug screening.
Keywords: IVIVC; bEnd.3 cell line; blood-brain barrier; mouse brain microvascular endothelial cells; primary cell culture.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures



Similar articles
-
Development of Human in vitro Brain-blood Barrier Model from Induced Pluripotent Stem Cell-derived Endothelial Cells to Predict the in vivo Permeability of Drugs.Neurosci Bull. 2019 Dec;35(6):996-1010. doi: 10.1007/s12264-019-00384-7. Epub 2019 May 11. Neurosci Bull. 2019. PMID: 31079318 Free PMC article.
-
A new blood-brain barrier model using primary rat brain endothelial cells, pericytes and astrocytes.Neurochem Int. 2009 Mar-Apr;54(3-4):253-63. doi: 10.1016/j.neuint.2008.12.002. Epub 2008 Dec 7. Neurochem Int. 2009. PMID: 19111869
-
In vitro models of the blood-brain barrier.Methods Mol Biol. 2014;1135:415-37. doi: 10.1007/978-1-4939-0320-7_34. Methods Mol Biol. 2014. PMID: 24510883
-
Human iPSC-Derived Blood-Brain Barrier Models: Valuable Tools for Preclinical Drug Discovery and Development?Curr Protoc Stem Cell Biol. 2020 Dec;55(1):e122. doi: 10.1002/cpsc.122. Curr Protoc Stem Cell Biol. 2020. PMID: 32956578 Review.
-
Co-Culture Models: Key Players in In Vitro Neurotoxicity, Neurodegeneration and BBB Modeling Studies.Biomedicines. 2024 Mar 12;12(3):626. doi: 10.3390/biomedicines12030626. Biomedicines. 2024. PMID: 38540242 Free PMC article. Review.
Cited by
-
Treatment of Alzheimer's Disease and Blood-Brain Barrier Drug Delivery.Pharmaceuticals (Basel). 2020 Nov 16;13(11):394. doi: 10.3390/ph13110394. Pharmaceuticals (Basel). 2020. PMID: 33207605 Free PMC article. Review.
-
Comparative studies between the murine immortalized brain endothelial cell line (bEnd.3) and induced pluripotent stem cell-derived human brain endothelial cells for paracellular transport.PLoS One. 2022 May 25;17(5):e0268860. doi: 10.1371/journal.pone.0268860. eCollection 2022. PLoS One. 2022. PMID: 35613139 Free PMC article.
-
Erratum: Puscas, I.; et al. IVIVC Assessment of Two Mouse Brain Endothelial Cell Models for Drug Screening. Pharmaceutics 2019, 11, 587.Pharmaceutics. 2020 Jun 4;12(6):514. doi: 10.3390/pharmaceutics12060514. Pharmaceutics. 2020. PMID: 32512770 Free PMC article.
-
Modeling blood-brain barrier pathology in cerebrovascular disease in vitro: current and future paradigms.Fluids Barriers CNS. 2020 Jul 16;17(1):44. doi: 10.1186/s12987-020-00202-7. Fluids Barriers CNS. 2020. PMID: 32677965 Free PMC article. Review.
-
Erythrocytes, a New Contributor to Age-Associated Loss of Blood-Brain Barrier Integrity.Adv Sci (Weinh). 2021 Oct;8(20):e2101912. doi: 10.1002/advs.202101912. Epub 2021 Aug 16. Adv Sci (Weinh). 2021. PMID: 34396716 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

